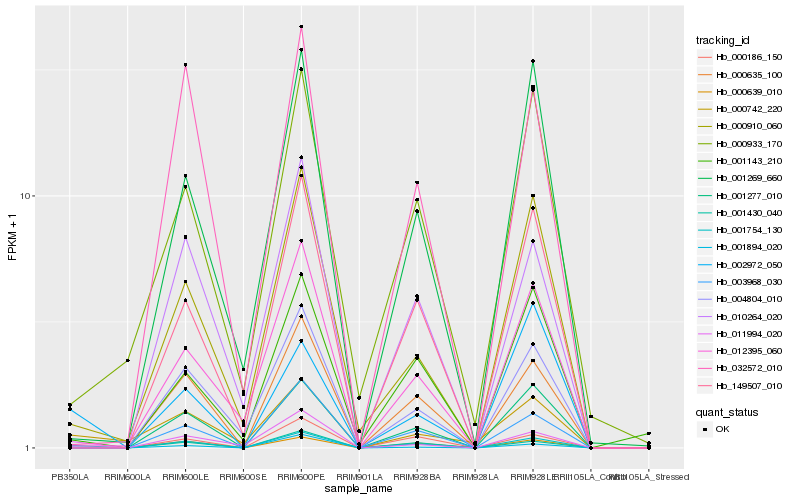
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001754_130 |
0.0 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 2 |
Hb_011994_020 |
0.176066598 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT5-like [Populus euphratica] |
| 3 |
Hb_000635_100 |
0.1859106374 |
- |
- |
- |
| 4 |
Hb_000910_060 |
0.1964830609 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 5 |
Hb_001277_010 |
0.1965776535 |
- |
- |
unknown [Populus trichocarpa] |
| 6 |
Hb_002972_050 |
0.1967023531 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000742_220 |
0.1967280188 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 8 |
Hb_003968_030 |
0.1992049749 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000639_010 |
0.2017808341 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_09727 [Jatropha curcas] |
| 10 |
Hb_001894_020 |
0.2032591587 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP630 [Jatropha curcas] |
| 11 |
Hb_149507_010 |
0.2050243521 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Malus domestica] |
| 12 |
Hb_001143_210 |
0.2061178393 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 13 |
Hb_001430_040 |
0.2067505928 |
- |
- |
gamma-tocopherol methyltransferase [Hevea brasiliensis] |
| 14 |
Hb_000186_150 |
0.2110160486 |
- |
- |
PREDICTED: light-induced protein, chloroplastic-like [Nicotiana sylvestris] |
| 15 |
Hb_000933_170 |
0.2137817699 |
- |
- |
PREDICTED: uncharacterized protein At5g01610 [Jatropha curcas] |
| 16 |
Hb_032572_010 |
0.2146191608 |
- |
- |
PREDICTED: phospholipase A2-alpha [Jatropha curcas] |
| 17 |
Hb_010264_020 |
0.214688198 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_004804_010 |
0.2147392807 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Populus euphratica] |
| 19 |
Hb_012395_060 |
0.214901256 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 2B [Jatropha curcas] |
| 20 |
Hb_001269_660 |
0.2164153618 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Jatropha curcas] |