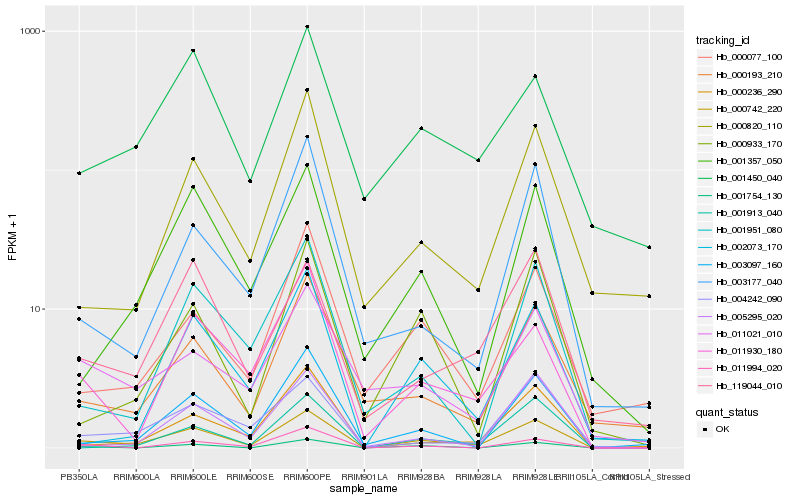
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000742_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 2 |
Hb_011930_180 |
0.1807485326 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004242_090 |
0.1818857009 |
- |
- |
hypothetical protein JCGZ_00767 [Jatropha curcas] |
| 4 |
Hb_119044_010 |
0.1887647867 |
- |
- |
PREDICTED: stomatal closure-related actin-binding protein 2-like [Jatropha curcas] |
| 5 |
Hb_000193_210 |
0.1892812052 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: galactoside 2-alpha-L-fucosyltransferase [Jatropha curcas] |
| 6 |
Hb_003097_160 |
0.1937145606 |
- |
- |
PREDICTED: F-box/WD repeat-containing protein sel-10 [Jatropha curcas] |
| 7 |
Hb_001357_050 |
0.1950104517 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12-like [Jatropha curcas] |
| 8 |
Hb_001754_130 |
0.1967280188 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 9 |
Hb_011994_020 |
0.1969142795 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT5-like [Populus euphratica] |
| 10 |
Hb_000236_290 |
0.197942487 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 11 |
Hb_002073_170 |
0.1985204447 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g27190 [Jatropha curcas] |
| 12 |
Hb_011021_010 |
0.2005975932 |
- |
- |
kinase, putative [Ricinus communis] |
| 13 |
Hb_000820_110 |
0.2015427614 |
- |
- |
UDP-XYL synthase 6 isoform 1 [Theobroma cacao] |
| 14 |
Hb_005295_020 |
0.201846709 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |
| 15 |
Hb_003177_040 |
0.2030411797 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 16 |
Hb_000933_170 |
0.2035749941 |
- |
- |
PREDICTED: uncharacterized protein At5g01610 [Jatropha curcas] |
| 17 |
Hb_001450_040 |
0.2037668724 |
- |
- |
PREDICTED: tubulin alpha-2 chain [Jatropha curcas] |
| 18 |
Hb_000077_100 |
0.2040105353 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001951_080 |
0.204507134 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 20 |
Hb_001913_040 |
0.205308294 |
- |
- |
conserved hypothetical protein [Ricinus communis] |