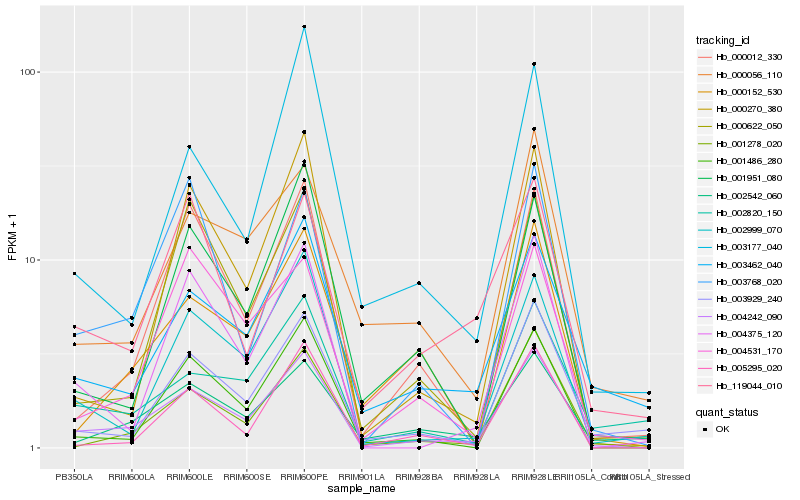
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004242_090 |
0.0 |
- |
- |
hypothetical protein JCGZ_00767 [Jatropha curcas] |
| 2 |
Hb_001278_020 |
0.140607897 |
- |
- |
sucrose synthase 6 [Hevea brasiliensis] |
| 3 |
Hb_002999_070 |
0.1506507735 |
- |
- |
glucose-methanol-choline (gmc) oxidoreductase, putative [Ricinus communis] |
| 4 |
Hb_001486_280 |
0.1511891204 |
- |
- |
hypothetical protein RCOM_1321910 [Ricinus communis] |
| 5 |
Hb_003929_240 |
0.1540269461 |
- |
- |
PREDICTED: uncharacterized protein LOC105643616 isoform X2 [Jatropha curcas] |
| 6 |
Hb_004375_120 |
0.1590359531 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 3-like [Jatropha curcas] |
| 7 |
Hb_003462_040 |
0.1594772222 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_002820_150 |
0.1628594542 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_000012_330 |
0.1653033305 |
- |
- |
pollen specific protein sf21, putative [Ricinus communis] |
| 10 |
Hb_000270_380 |
0.166930631 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 11 |
Hb_005295_020 |
0.1691573054 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |
| 12 |
Hb_000056_110 |
0.1718227459 |
- |
- |
PREDICTED: uncharacterized protein LOC105630405 [Jatropha curcas] |
| 13 |
Hb_002542_060 |
0.1718315215 |
- |
- |
PREDICTED: wee1-like protein kinase [Jatropha curcas] |
| 14 |
Hb_000622_050 |
0.1727500472 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003768_020 |
0.1760788703 |
- |
- |
PREDICTED: uncharacterized protein LOC105644157 [Jatropha curcas] |
| 16 |
Hb_001951_080 |
0.1781887949 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 17 |
Hb_000152_530 |
0.1781981897 |
- |
- |
hypothetical protein SELMODRAFT_231485 [Selaginella moellendorffii] |
| 18 |
Hb_003177_040 |
0.1790638028 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 19 |
Hb_004531_170 |
0.1796296872 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 20 |
Hb_119044_010 |
0.1802125294 |
- |
- |
PREDICTED: stomatal closure-related actin-binding protein 2-like [Jatropha curcas] |