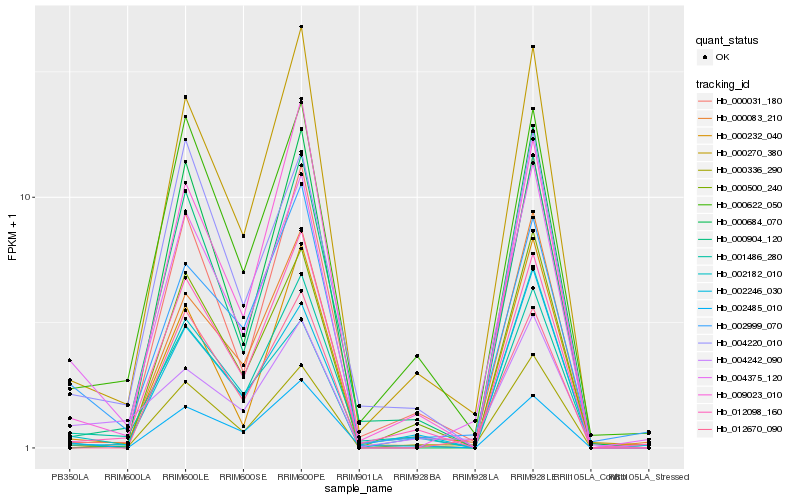
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004375_120 |
0.0 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 3-like [Jatropha curcas] |
| 2 |
Hb_000336_290 |
0.1187276792 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 3 |
Hb_002999_070 |
0.1235165473 |
- |
- |
glucose-methanol-choline (gmc) oxidoreductase, putative [Ricinus communis] |
| 4 |
Hb_000270_380 |
0.131768724 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 5 |
Hb_001486_280 |
0.1414171924 |
- |
- |
hypothetical protein RCOM_1321910 [Ricinus communis] |
| 6 |
Hb_000622_050 |
0.1535533068 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002182_010 |
0.1547876094 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase-like [Jatropha curcas] |
| 8 |
Hb_004220_010 |
0.1581967072 |
- |
- |
PREDICTED: uncharacterized protein LOC105642366 [Jatropha curcas] |
| 9 |
Hb_004242_090 |
0.1590359531 |
- |
- |
hypothetical protein JCGZ_00767 [Jatropha curcas] |
| 10 |
Hb_012098_160 |
0.1598158298 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 11 |
Hb_002485_010 |
0.1604808457 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 12 |
Hb_000904_120 |
0.1618567134 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000232_040 |
0.1640216697 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X2 [Populus euphratica] |
| 14 |
Hb_002246_030 |
0.165265054 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 15 |
Hb_000031_180 |
0.1662356361 |
- |
- |
sarcosine oxidase, putative [Ricinus communis] |
| 16 |
Hb_000083_210 |
0.166853291 |
- |
- |
Major facilitator superfamily protein, putative [Theobroma cacao] |
| 17 |
Hb_012670_090 |
0.1670461565 |
- |
- |
GDSL-motif lipase/hydrolase family protein [Populus trichocarpa] |
| 18 |
Hb_009023_010 |
0.1675301792 |
- |
- |
glycosyl hydrolase family 31 family protein [Populus trichocarpa] |
| 19 |
Hb_000500_240 |
0.1681394166 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000684_070 |
0.1686787687 |
- |
- |
PREDICTED: uncharacterized protein LOC105642198 isoform X1 [Jatropha curcas] |