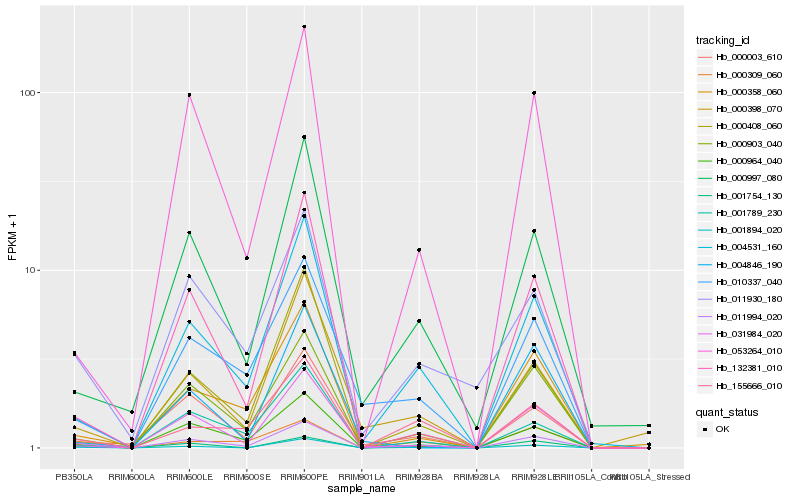
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011994_020 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT5-like [Populus euphratica] |
| 2 |
Hb_001894_020 |
0.155599121 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP630 [Jatropha curcas] |
| 3 |
Hb_011930_180 |
0.1626095599 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000903_040 |
0.1729004993 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |
| 5 |
Hb_000964_040 |
0.175507274 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER isoform X2 [Jatropha curcas] |
| 6 |
Hb_001754_130 |
0.176066598 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 7 |
Hb_000309_060 |
0.1780601643 |
- |
- |
hypothetical protein JCGZ_09609 [Jatropha curcas] |
| 8 |
Hb_000358_060 |
0.1780745121 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 9 |
Hb_053264_010 |
0.1800775151 |
- |
- |
beta glucosidase [Hevea brasiliensis] |
| 10 |
Hb_000003_610 |
0.1816374839 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase [Jatropha curcas] |
| 11 |
Hb_155666_010 |
0.1850145856 |
- |
- |
hypothetical protein JCGZ_01214 [Jatropha curcas] |
| 12 |
Hb_031984_020 |
0.186057744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_132381_010 |
0.1866168509 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 14 |
Hb_000997_080 |
0.1868944724 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000408_060 |
0.1879372509 |
- |
- |
hypothetical protein POPTR_0018s11360g [Populus trichocarpa] |
| 16 |
Hb_010337_040 |
0.1923888973 |
- |
- |
- |
| 17 |
Hb_004846_190 |
0.1924668329 |
- |
- |
hypothetical protein POPTR_0001s03430g [Populus trichocarpa] |
| 18 |
Hb_001789_230 |
0.1933097399 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000398_070 |
0.1951035744 |
- |
- |
hypothetical protein VITISV_011880 [Vitis vinifera] |
| 20 |
Hb_004531_160 |
0.1951479295 |
- |
- |
PREDICTED: transcription factor bHLH68-like isoform X1 [Jatropha curcas] |