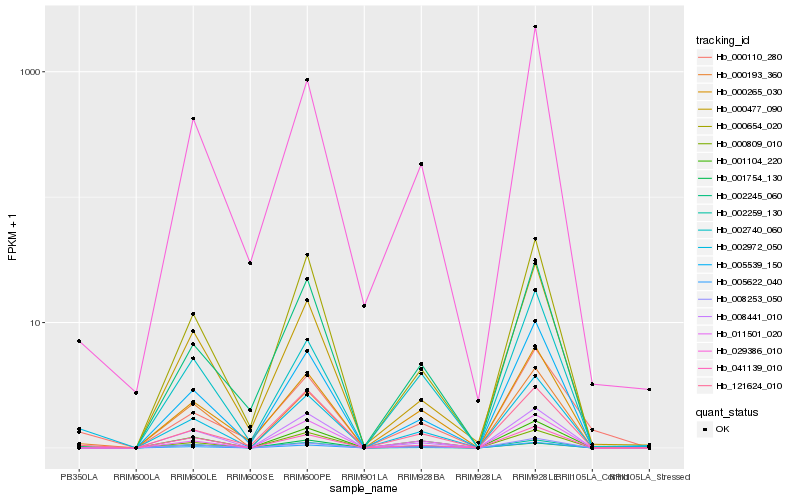
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002972_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002259_130 |
0.1748944288 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_121624_010 |
0.1834768336 |
- |
- |
PREDICTED: uncharacterized protein LOC105634410 [Jatropha curcas] |
| 4 |
Hb_000110_280 |
0.1854763885 |
- |
- |
PREDICTED: uncharacterized protein LOC105642012, partial [Jatropha curcas] |
| 5 |
Hb_001754_130 |
0.1967023531 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 6 |
Hb_008253_050 |
0.2000546737 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0001s36220g [Populus trichocarpa] |
| 7 |
Hb_001104_220 |
0.2014328951 |
- |
- |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 8 |
Hb_005622_040 |
0.2033538553 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 9 |
Hb_008441_010 |
0.2072433901 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 10 |
Hb_000193_360 |
0.2073563007 |
- |
- |
PREDICTED: uncharacterized protein At5g01610-like [Jatropha curcas] |
| 11 |
Hb_005539_150 |
0.207479555 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 33 [Jatropha curcas] |
| 12 |
Hb_041139_010 |
0.2105038018 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 13 |
Hb_000654_020 |
0.2112466585 |
- |
- |
hypothetical protein CICLE_v10006262mg [Citrus clementina] |
| 14 |
Hb_011501_020 |
0.2123579908 |
- |
- |
PREDICTED: uncharacterized protein LOC105648929 [Jatropha curcas] |
| 15 |
Hb_029386_010 |
0.2151745342 |
- |
- |
RALF-LIKE 27 family protein [Populus trichocarpa] |
| 16 |
Hb_000265_030 |
0.2160067718 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 17 |
Hb_002245_060 |
0.2196009251 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000809_010 |
0.2201417185 |
- |
- |
hypothetical protein JCGZ_12342 [Jatropha curcas] |
| 19 |
Hb_002740_060 |
0.2221312191 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 20 |
Hb_000477_090 |
0.2231294813 |
- |
- |
PREDICTED: uncharacterized protein LOC105639302 [Jatropha curcas] |