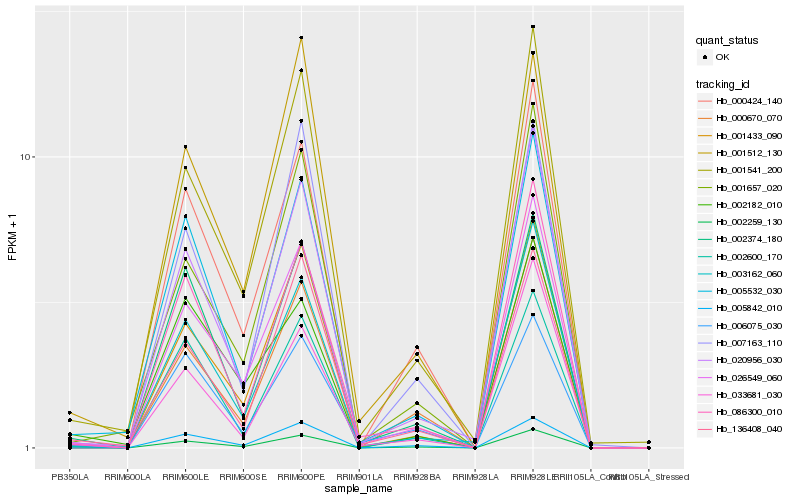
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002259_130 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_001541_200 |
0.112315356 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 3 |
Hb_026549_060 |
0.1124839196 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 4 |
Hb_136408_040 |
0.1165366344 |
- |
- |
PREDICTED: protein NDR1 [Jatropha curcas] |
| 5 |
Hb_001433_090 |
0.1203926814 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 6 |
Hb_005532_030 |
0.1216089844 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_002374_180 |
0.1223559302 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 8 |
Hb_020956_030 |
0.1246880856 |
- |
- |
PREDICTED: swi5-dependent recombination DNA repair protein 1 homolog [Jatropha curcas] |
| 9 |
Hb_002600_170 |
0.1285364933 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005842_010 |
0.1310255805 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 11 |
Hb_000424_140 |
0.1320387756 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_086300_010 |
0.1329554851 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 13 |
Hb_000670_070 |
0.1331200129 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 14 |
Hb_001657_020 |
0.1350561497 |
- |
- |
heat shock family protein [Populus trichocarpa] |
| 15 |
Hb_033681_030 |
0.1357249972 |
desease resistance |
Gene Name: NB-ARC |
Leucine-rich repeat containing protein [Theobroma cacao] |
| 16 |
Hb_007163_110 |
0.1371640314 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 17 |
Hb_006075_030 |
0.137864713 |
- |
- |
PREDICTED: uncharacterized protein LOC105641890 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003162_060 |
0.1383182355 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 19 |
Hb_001512_130 |
0.139130684 |
- |
- |
PREDICTED: uncharacterized protein LOC105633633 [Jatropha curcas] |
| 20 |
Hb_002182_010 |
0.1405956538 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase-like [Jatropha curcas] |