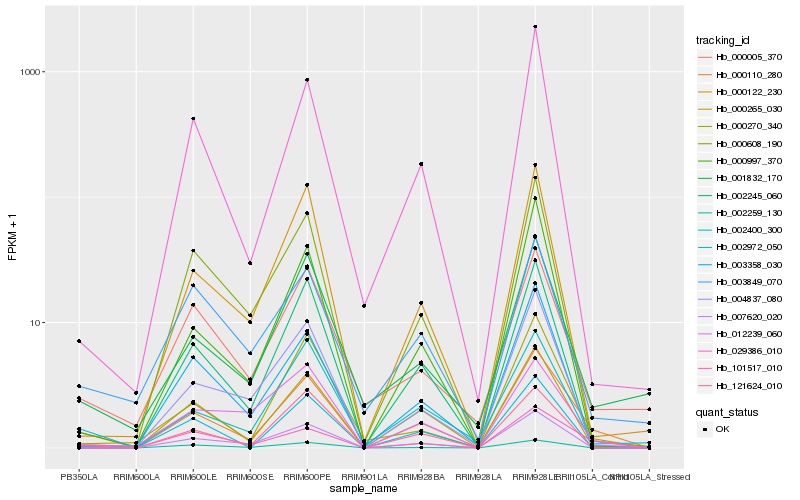
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000110_280 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642012, partial [Jatropha curcas] |
| 2 |
Hb_101517_010 |
0.1786061379 |
- |
- |
PREDICTED: uncharacterized protein LOC105629574 [Jatropha curcas] |
| 3 |
Hb_001832_170 |
0.182778518 |
- |
- |
PREDICTED: beta-galactosidase-like [Jatropha curcas] |
| 4 |
Hb_002972_050 |
0.1854763885 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_121624_010 |
0.1879483068 |
- |
- |
PREDICTED: uncharacterized protein LOC105634410 [Jatropha curcas] |
| 6 |
Hb_002259_130 |
0.1898418105 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_000265_030 |
0.1899040857 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 8 |
Hb_000270_340 |
0.1904621286 |
- |
- |
hypothetical protein JCGZ_11546 [Jatropha curcas] |
| 9 |
Hb_000005_370 |
0.1919136617 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105634024 [Jatropha curcas] |
| 10 |
Hb_029386_010 |
0.1962861112 |
- |
- |
RALF-LIKE 27 family protein [Populus trichocarpa] |
| 11 |
Hb_002245_060 |
0.1980232822 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000122_230 |
0.1988200216 |
- |
- |
laccase, putative [Ricinus communis] |
| 13 |
Hb_000997_370 |
0.1989984169 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Jatropha curcas] |
| 14 |
Hb_002400_300 |
0.1995745959 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 15 |
Hb_003849_070 |
0.2013067526 |
- |
- |
PREDICTED: uncharacterized protein LOC105644690 [Jatropha curcas] |
| 16 |
Hb_007620_020 |
0.2014217661 |
- |
- |
PREDICTED: cationic amino acid transporter 7, chloroplastic-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_004837_080 |
0.2019735026 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46 [Jatropha curcas] |
| 18 |
Hb_012239_060 |
0.2030739805 |
- |
- |
PREDICTED: nudix hydrolase 18, mitochondrial [Jatropha curcas] |
| 19 |
Hb_003358_030 |
0.2031453543 |
- |
- |
ankyrin-kinase, putative [Ricinus communis] |
| 20 |
Hb_000608_190 |
0.2037578009 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |