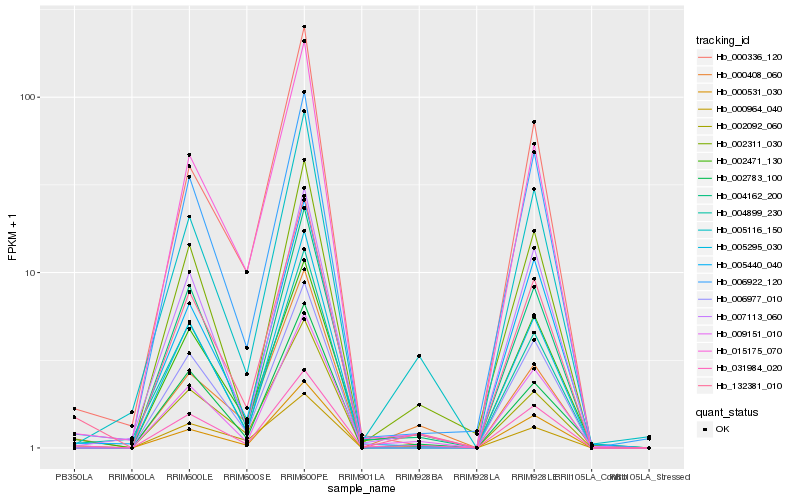
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_132381_010 |
0.0 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 2 |
Hb_031984_020 |
0.0756518871 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_015175_070 |
0.0924950527 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 4 |
Hb_000531_030 |
0.0954705041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002471_130 |
0.0961863328 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 3 [Jatropha curcas] |
| 6 |
Hb_005295_030 |
0.0972108941 |
- |
- |
PREDICTED: uncharacterized protein LOC105645488 [Jatropha curcas] |
| 7 |
Hb_004899_230 |
0.0994802351 |
- |
- |
Indole-3-acetic acid-induced protein ARG7, putative [Ricinus communis] |
| 8 |
Hb_004162_200 |
0.101200043 |
- |
- |
PREDICTED: uncharacterized protein At1g08160 [Jatropha curcas] |
| 9 |
Hb_006922_120 |
0.1025564108 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 10 |
Hb_006977_010 |
0.1033126041 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_002092_060 |
0.1049068323 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000336_120 |
0.1049529663 |
- |
- |
PREDICTED: mavicyanin [Populus euphratica] |
| 13 |
Hb_005440_040 |
0.1063305974 |
- |
- |
PREDICTED: microtubule-associated protein 70-5 [Jatropha curcas] |
| 14 |
Hb_005116_150 |
0.1080341682 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_007113_060 |
0.1085829986 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 16 |
Hb_000408_060 |
0.1094587985 |
- |
- |
hypothetical protein POPTR_0018s11360g [Populus trichocarpa] |
| 17 |
Hb_009151_010 |
0.1098756873 |
- |
- |
Peroxidase 3 [Morus notabilis] |
| 18 |
Hb_002783_100 |
0.1100969809 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 19 |
Hb_002311_030 |
0.1113433845 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000964_040 |
0.1123254038 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER isoform X2 [Jatropha curcas] |