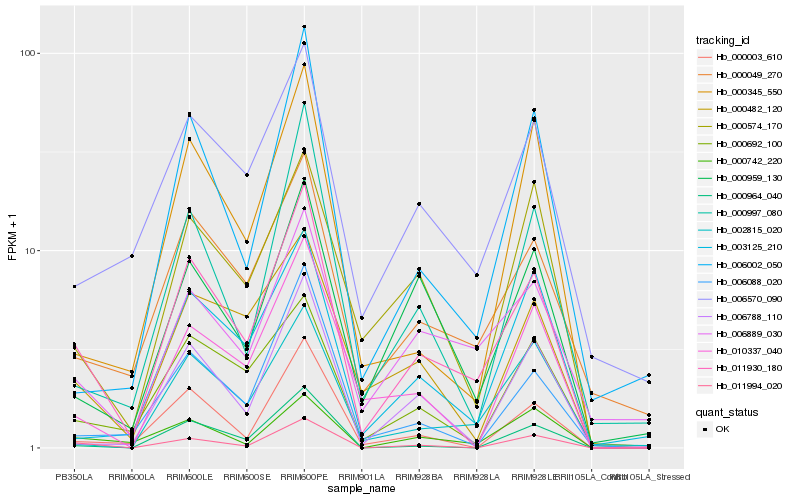
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011930_180 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000049_270 |
0.1497405178 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_006889_030 |
0.1606953888 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 4 |
Hb_011994_020 |
0.1626095599 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT5-like [Populus euphratica] |
| 5 |
Hb_006570_090 |
0.1674418693 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 6 |
Hb_000692_100 |
0.1703751226 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 7 |
Hb_000482_120 |
0.1745717054 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |
| 8 |
Hb_000003_610 |
0.1748513097 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase [Jatropha curcas] |
| 9 |
Hb_000959_130 |
0.1762959185 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 10 |
Hb_000742_220 |
0.1807485326 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 11 |
Hb_000997_080 |
0.182414678 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_006088_020 |
0.1825401205 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 13 |
Hb_006788_110 |
0.183071714 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 14 |
Hb_000574_170 |
0.1832247993 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump-like [Jatropha curcas] |
| 15 |
Hb_006002_050 |
0.1834113747 |
- |
- |
PREDICTED: uncharacterized protein LOC105638083 [Jatropha curcas] |
| 16 |
Hb_002815_020 |
0.1841217082 |
transcription factor |
TF Family: G2-like |
Homeodomain-like superfamily protein isoform 2 [Theobroma cacao] |
| 17 |
Hb_010337_040 |
0.1873091237 |
- |
- |
- |
| 18 |
Hb_003125_210 |
0.187866491 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 19 |
Hb_000345_550 |
0.1881902317 |
- |
- |
PREDICTED: uncharacterized protein LOC105629456 [Jatropha curcas] |
| 20 |
Hb_000964_040 |
0.1892933291 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER isoform X2 [Jatropha curcas] |