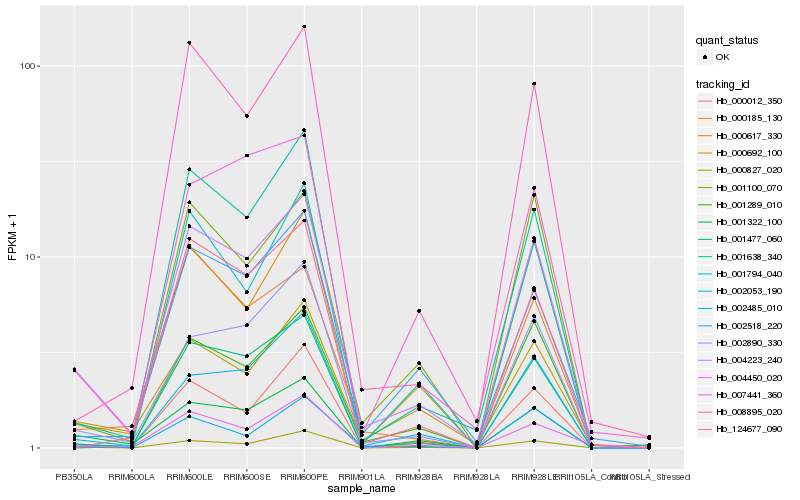
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004223_240 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635369 [Jatropha curcas] |
| 2 |
Hb_000617_330 |
0.1196393472 |
- |
- |
PREDICTED: uncharacterized protein LOC105647507 [Jatropha curcas] |
| 3 |
Hb_002890_330 |
0.1222943678 |
- |
- |
caspase, putative [Ricinus communis] |
| 4 |
Hb_001289_010 |
0.122952757 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Vitis vinifera] |
| 5 |
Hb_001638_340 |
0.1244186974 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000692_100 |
0.1248657314 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 7 |
Hb_001322_100 |
0.1268831499 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase [Vitis vinifera] |
| 8 |
Hb_000827_020 |
0.1316070888 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_001477_060 |
0.1365680633 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |
| 10 |
Hb_008895_020 |
0.1377224114 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 11 |
Hb_001100_070 |
0.1378472101 |
- |
- |
PREDICTED: ferredoxin--nitrite reductase, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_002518_220 |
0.1392285975 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000012_350 |
0.1416119026 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein ATH1 [Jatropha curcas] |
| 14 |
Hb_002053_190 |
0.1419336382 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004450_020 |
0.1427830305 |
- |
- |
serine-threonine protein kinase, putative [Ricinus communis] |
| 16 |
Hb_002485_010 |
0.1446688246 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 17 |
Hb_001794_040 |
0.1455636903 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 18 |
Hb_124677_090 |
0.1456197955 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000185_130 |
0.1464377827 |
- |
- |
UBX domain-containing protein 8-B, putative [Ricinus communis] |
| 20 |
Hb_007441_360 |
0.1471957675 |
- |
- |
PREDICTED: uncharacterized protein LOC105643270 [Jatropha curcas] |