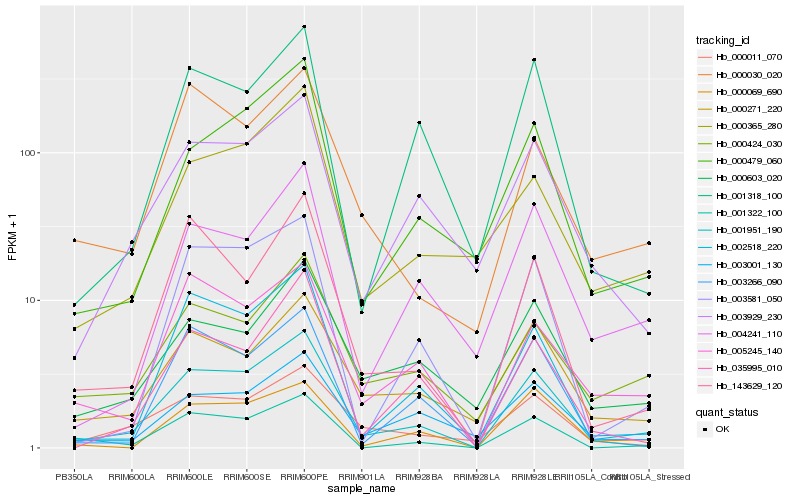
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001951_190 |
0.0 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 3-like isoform X2 [Populus euphratica] |
| 2 |
Hb_005245_140 |
0.1427879184 |
- |
- |
PREDICTED: ribonuclease 2 [Jatropha curcas] |
| 3 |
Hb_000479_060 |
0.1442167107 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1 [Vitis vinifera] |
| 4 |
Hb_003266_090 |
0.1476784124 |
- |
- |
PREDICTED: basic 7S globulin 2-like [Jatropha curcas] |
| 5 |
Hb_003581_050 |
0.1489680272 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-54 [Jatropha curcas] |
| 6 |
Hb_000424_030 |
0.1511526286 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003929_230 |
0.1522306787 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 5 [Nelumbo nucifera] |
| 8 |
Hb_003001_130 |
0.1526337746 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_143629_120 |
0.1531291712 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 10 |
Hb_000271_220 |
0.1537854722 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 11 |
Hb_001318_100 |
0.1552156461 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 12 |
Hb_004241_110 |
0.1552654939 |
- |
- |
Minor allergen Alt a, putative [Ricinus communis] |
| 13 |
Hb_000069_690 |
0.1559993092 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF6 [Jatropha curcas] |
| 14 |
Hb_002518_220 |
0.1575662908 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 15 |
Hb_035995_010 |
0.1579510446 |
- |
- |
- |
| 16 |
Hb_000365_280 |
0.1579917601 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 17 |
Hb_000603_020 |
0.1593554867 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000011_070 |
0.1593906232 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 19 |
Hb_001322_100 |
0.15967804 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase [Vitis vinifera] |
| 20 |
Hb_000030_020 |
0.1606792598 |
- |
- |
PREDICTED: uncharacterized protein LOC105123549 [Populus euphratica] |