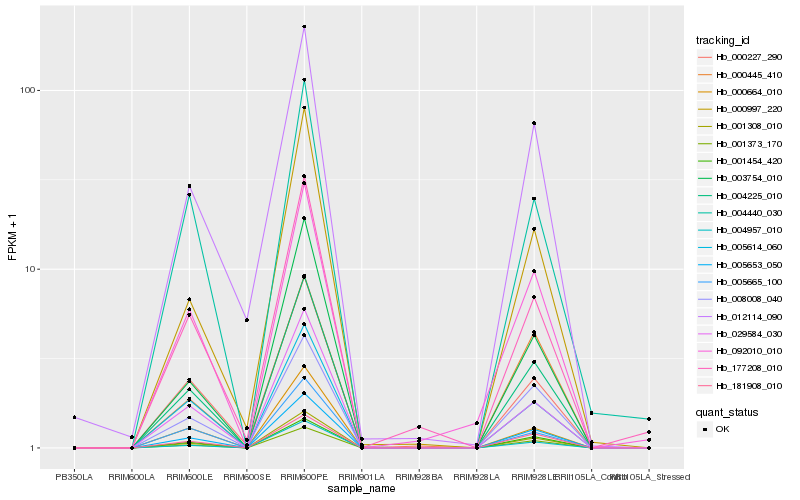
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004225_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_005653_050 |
0.008913643 |
- |
- |
PREDICTED: CMP-N-acetylneuraminate-beta-1,4-galactoside alpha-2,3-sialyltransferase [Jatropha curcas] |
| 3 |
Hb_001308_010 |
0.0397205146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001373_170 |
0.0445831219 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 9 [Jatropha curcas] |
| 5 |
Hb_001454_420 |
0.0521877524 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 6 |
Hb_029584_030 |
0.0658242767 |
- |
- |
hypothetical protein JCGZ_07310 [Jatropha curcas] |
| 7 |
Hb_181908_010 |
0.0689142851 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C3 [Jatropha curcas] |
| 8 |
Hb_004957_010 |
0.0708134914 |
- |
- |
PREDICTED: probable aquaporin NIP-type [Prunus mume] |
| 9 |
Hb_008008_040 |
0.0715880821 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 10 |
Hb_005614_060 |
0.074654324 |
- |
- |
Uncharacterized protein TCM_019750 [Theobroma cacao] |
| 11 |
Hb_000664_010 |
0.0754927987 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 4-like [Jatropha curcas] |
| 12 |
Hb_000227_290 |
0.0815634719 |
- |
- |
PREDICTED: uncharacterized protein LOC105631957 [Jatropha curcas] |
| 13 |
Hb_003754_010 |
0.0839685218 |
- |
- |
hypothetical protein POPTR_0019s09950g [Populus trichocarpa] |
| 14 |
Hb_092010_010 |
0.0896355678 |
- |
- |
cysteine proteinase inhibitor 5-like [Cucumis sativus] |
| 15 |
Hb_000997_220 |
0.089696344 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA14-like [Jatropha curcas] |
| 16 |
Hb_004440_030 |
0.0929317139 |
transcription factor |
TF Family: MYB-related |
hypothetical protein POPTR_0005s12390g [Populus trichocarpa] |
| 17 |
Hb_012114_090 |
0.0932509402 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 18 |
Hb_005665_100 |
0.094382492 |
- |
- |
PREDICTED: RING-H2 finger protein ATL63-like [Populus euphratica] |
| 19 |
Hb_177208_010 |
0.0982194251 |
transcription factor |
TF Family: MIKC |
MADS-box transcription factor, partial [Jatropha sp. CONN 198700232] |
| 20 |
Hb_000445_410 |
0.1014122249 |
- |
- |
hypothetical protein EUGRSUZ_F02776 [Eucalyptus grandis] |