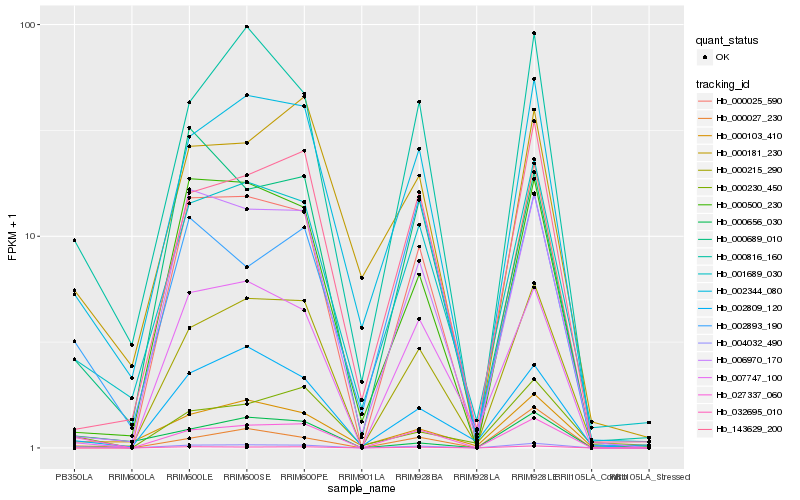
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004032_490 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC100853401 [Vitis vinifera] |
| 2 |
Hb_027337_060 |
0.0640249651 |
- |
- |
- |
| 3 |
Hb_002344_080 |
0.1592538523 |
- |
- |
PREDICTED: protein YLS2-like [Populus euphratica] |
| 4 |
Hb_000816_160 |
0.1626427508 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 5 |
Hb_001689_030 |
0.1650801908 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |
| 6 |
Hb_000103_410 |
0.1928724682 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 7 |
Hb_000230_450 |
0.1939304756 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000025_590 |
0.2003791111 |
- |
- |
PREDICTED: cytochrome P450 94A1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000656_030 |
0.2020710155 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002893_190 |
0.2037703893 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_000181_230 |
0.2051484283 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 12 |
Hb_002809_120 |
0.2059838483 |
- |
- |
Uncharacterized protein TCM_031281 [Theobroma cacao] |
| 13 |
Hb_006970_170 |
0.2087458156 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_000689_010 |
0.2087888568 |
desease resistance |
Gene Name: LRR_8 |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 15 |
Hb_000215_290 |
0.2089615537 |
transcription factor |
TF Family: NAC |
PREDICTED: protein BEARSKIN2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_032695_010 |
0.2091095192 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2 [Vitis vinifera] |
| 17 |
Hb_007747_100 |
0.2106601623 |
- |
- |
PREDICTED: probable F-box protein At1g60180 [Jatropha curcas] |
| 18 |
Hb_000027_230 |
0.2115094364 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 19 |
Hb_000500_230 |
0.2121640526 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 20 |
Hb_143629_200 |
0.2123590566 |
- |
- |
hypothetical protein POPTR_0013s00300g [Populus trichocarpa] |