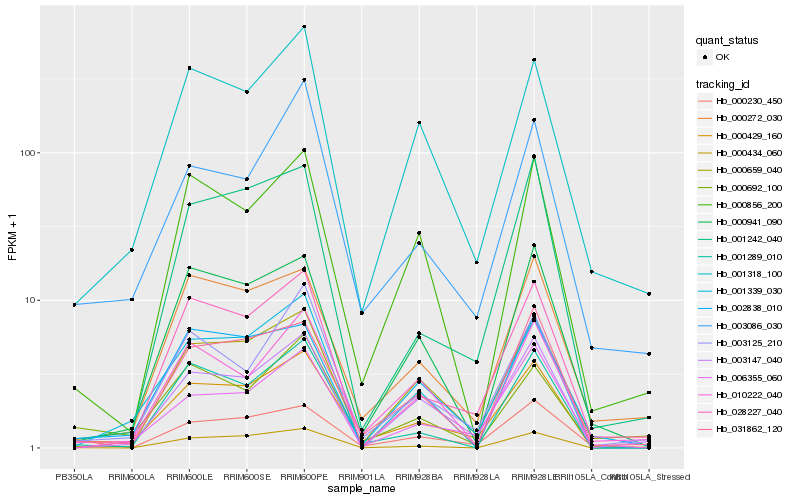
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000429_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645177 [Jatropha curcas] |
| 2 |
Hb_001318_100 |
0.1214842761 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 3 |
Hb_003125_210 |
0.1259373068 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 4 |
Hb_031862_120 |
0.1259874545 |
- |
- |
PREDICTED: root phototropism protein 3 [Jatropha curcas] |
| 5 |
Hb_028227_040 |
0.1295971577 |
- |
- |
PREDICTED: uncharacterized protein LOC105645005 [Jatropha curcas] |
| 6 |
Hb_003147_040 |
0.130244453 |
- |
- |
PREDICTED: probable calcium-binding protein CML18 [Jatropha curcas] |
| 7 |
Hb_000659_040 |
0.1307922245 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000692_100 |
0.1358263812 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 9 |
Hb_000272_030 |
0.1364857791 |
- |
- |
PREDICTED: WD repeat-containing protein 76 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002838_010 |
0.1384693622 |
- |
- |
AGPase large subunit protein [Hevea brasiliensis] |
| 11 |
Hb_001339_030 |
0.1385211043 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 11-like [Jatropha curcas] |
| 12 |
Hb_001289_010 |
0.1390160385 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Vitis vinifera] |
| 13 |
Hb_006355_060 |
0.1391300432 |
- |
- |
PREDICTED: transcription factor PIF1-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_003086_030 |
0.140297783 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 15 |
Hb_000434_060 |
0.1423092757 |
desease resistance |
Gene Name: hmm |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 16 |
Hb_000856_200 |
0.1438018209 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 17 |
Hb_000941_090 |
0.1448742796 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 18 |
Hb_001242_040 |
0.145202766 |
- |
- |
PREDICTED: myb-related transcription factor, partner of profilin-like [Populus euphratica] |
| 19 |
Hb_000230_450 |
0.1456206058 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_010222_040 |
0.1466646336 |
- |
- |
PREDICTED: kinesin-2 [Jatropha curcas] |