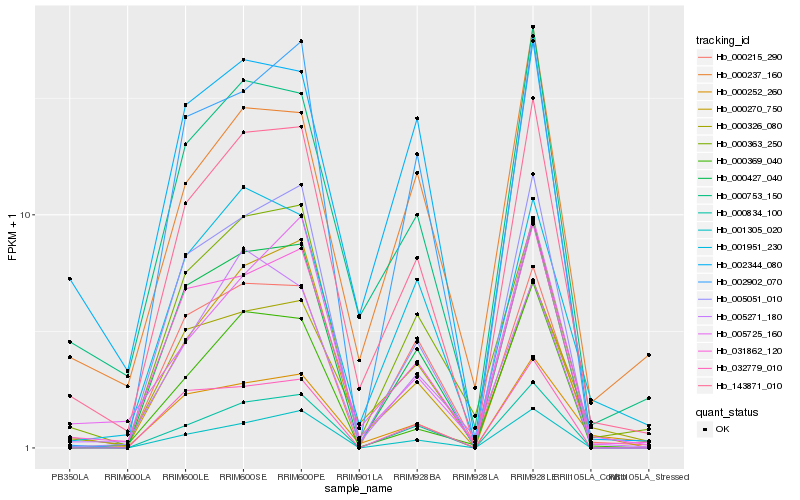
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_143871_010 |
0.0 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 2 |
Hb_031862_120 |
0.0787053042 |
- |
- |
PREDICTED: root phototropism protein 3 [Jatropha curcas] |
| 3 |
Hb_000753_150 |
0.0831979546 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.4 [Jatropha curcas] |
| 4 |
Hb_000834_100 |
0.1072700289 |
- |
- |
PREDICTED: uncharacterized protein LOC105178355 [Sesamum indicum] |
| 5 |
Hb_000427_040 |
0.1074135769 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 6 |
Hb_005051_010 |
0.108794598 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_000326_080 |
0.1134386688 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 8 |
Hb_000363_250 |
0.124705959 |
- |
- |
PREDICTED: pirin-like protein [Malus domestica] |
| 9 |
Hb_001305_020 |
0.1253514615 |
- |
- |
PREDICTED: uncharacterized protein LOC102623247 [Citrus sinensis] |
| 10 |
Hb_000215_290 |
0.1255808894 |
transcription factor |
TF Family: NAC |
PREDICTED: protein BEARSKIN2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001951_230 |
0.1313568045 |
- |
- |
PREDICTED: phytochrome E isoform X2 [Jatropha curcas] |
| 12 |
Hb_000270_750 |
0.13383698 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46-like [Populus euphratica] |
| 13 |
Hb_005725_160 |
0.1344095378 |
- |
- |
PREDICTED: uncharacterized protein LOC105630811 [Jatropha curcas] |
| 14 |
Hb_002902_070 |
0.1349412604 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g67385 [Jatropha curcas] |
| 15 |
Hb_032779_010 |
0.1367226597 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_002344_080 |
0.1368450483 |
- |
- |
PREDICTED: protein YLS2-like [Populus euphratica] |
| 17 |
Hb_000369_040 |
0.1390533551 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 18 |
Hb_000252_260 |
0.1395749802 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 19 |
Hb_005271_180 |
0.1402507654 |
- |
- |
Leucine-rich repeat receptor protein kinase EXS precursor, putative [Ricinus communis] |
| 20 |
Hb_000237_160 |
0.1417292876 |
- |
- |
BEL1-like homeodomain protein 1 [Morus notabilis] |