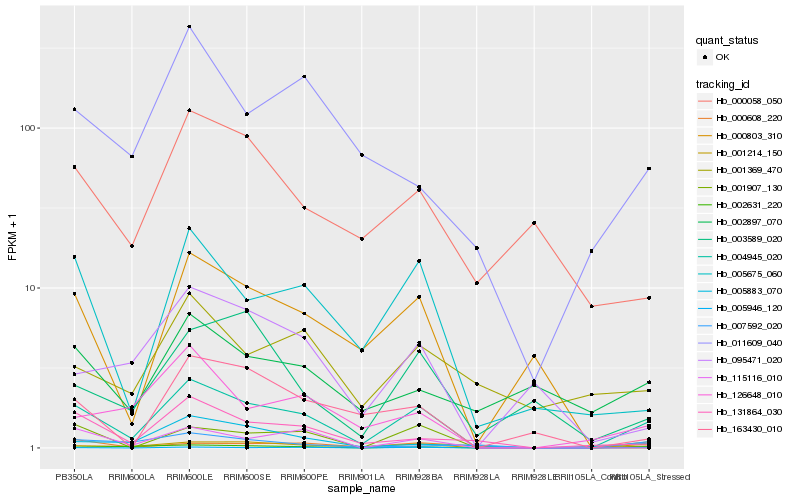
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004945_020 |
0.0 |
- |
- |
PREDICTED: probable calcium-binding protein CML30 [Vitis vinifera] |
| 2 |
Hb_001214_150 |
0.2168111579 |
- |
- |
PREDICTED: uncharacterized protein LOC102624288 [Citrus sinensis] |
| 3 |
Hb_005946_120 |
0.2262755348 |
- |
- |
peptidase, putative [Ricinus communis] |
| 4 |
Hb_002631_220 |
0.2361080254 |
- |
- |
Patellin-4, putative [Ricinus communis] |
| 5 |
Hb_005675_060 |
0.2362552562 |
- |
- |
PREDICTED: protein NIM1-INTERACTING 2 [Jatropha curcas] |
| 6 |
Hb_131864_030 |
0.2587402538 |
transcription factor |
TF Family: C2H2 |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_126648_010 |
0.2603601913 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 33 homolog [Jatropha curcas] |
| 8 |
Hb_000608_220 |
0.2657533004 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 9 |
Hb_163430_010 |
0.2746290338 |
- |
- |
Polyneuridine-aldehyde esterase precursor, putative [Ricinus communis] |
| 10 |
Hb_005883_070 |
0.2756141838 |
- |
- |
PREDICTED: probable myosin-binding protein 5 [Jatropha curcas] |
| 11 |
Hb_011609_040 |
0.2778307733 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 40 [Jatropha curcas] |
| 12 |
Hb_115116_010 |
0.2788117938 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 13 |
Hb_095471_020 |
0.2820655203 |
- |
- |
- |
| 14 |
Hb_007592_020 |
0.2844260107 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 15 |
Hb_000803_310 |
0.2855030309 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105648332 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003589_020 |
0.2872175555 |
- |
- |
- |
| 17 |
Hb_000058_050 |
0.2898122244 |
- |
- |
PREDICTED: uncharacterized protein LOC105640877 [Jatropha curcas] |
| 18 |
Hb_001907_130 |
0.2901202097 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002897_070 |
0.2938686609 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 20 |
Hb_001369_470 |
0.2962789043 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKS1 [Jatropha curcas] |