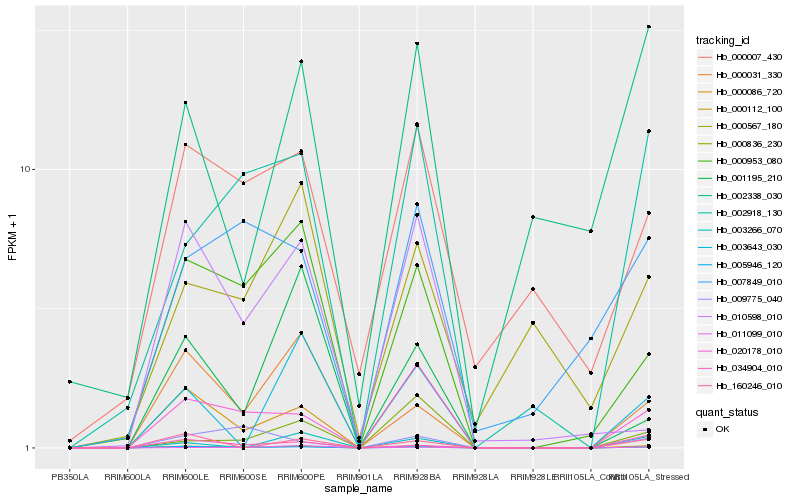
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000112_100 |
0.0 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 2 |
Hb_020178_010 |
0.139821847 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein A-like [Jatropha curcas] |
| 3 |
Hb_000953_080 |
0.2029413972 |
- |
- |
PREDICTED: PAN domain-containing protein At5g03700 [Jatropha curcas] |
| 4 |
Hb_002918_130 |
0.209562647 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 12 [Jatropha curcas] |
| 5 |
Hb_005946_120 |
0.2320256213 |
- |
- |
peptidase, putative [Ricinus communis] |
| 6 |
Hb_000031_330 |
0.240999458 |
transcription factor |
TF Family: Orphans |
hypothetical protein POPTR_0157s00200g, partial [Populus trichocarpa] |
| 7 |
Hb_000007_430 |
0.2545514423 |
- |
- |
hypothetical protein POPTR_0010s14280g [Populus trichocarpa] |
| 8 |
Hb_007849_010 |
0.2546909338 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 9 |
Hb_000836_230 |
0.2643773371 |
- |
- |
Uncharacterized protein TCM_041879 [Theobroma cacao] |
| 10 |
Hb_160246_010 |
0.2672915841 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 11 |
Hb_011099_010 |
0.2707943837 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 12 |
Hb_034904_010 |
0.2728700859 |
- |
- |
PREDICTED: uncharacterized protein LOC105775231 [Gossypium raimondii] |
| 13 |
Hb_003643_030 |
0.280139962 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Populus euphratica] |
| 14 |
Hb_010598_010 |
0.285985059 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 15 |
Hb_009775_040 |
0.2862662826 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Vitis vinifera] |
| 16 |
Hb_000567_180 |
0.2922959123 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.5 [Jatropha curcas] |
| 17 |
Hb_003266_070 |
0.2934734609 |
- |
- |
- |
| 18 |
Hb_000086_720 |
0.2960465653 |
- |
- |
leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 19 |
Hb_002338_030 |
0.2963898712 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P1-like [Tarenaya hassleriana] |
| 20 |
Hb_001195_210 |
0.2992093856 |
- |
- |
- |