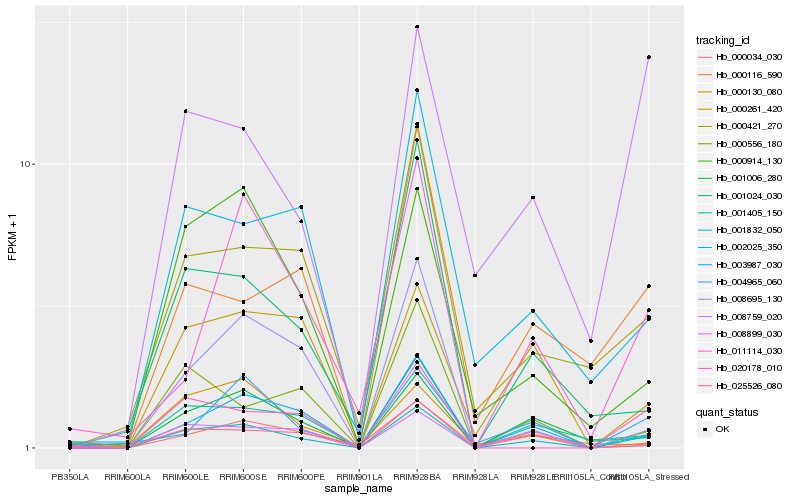
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001832_050 |
0.0 |
- |
- |
PREDICTED: cytokinin dehydrogenase 3 [Jatropha curcas] |
| 2 |
Hb_001405_150 |
0.1839984929 |
- |
- |
PREDICTED: protein At-4/1 [Jatropha curcas] |
| 3 |
Hb_003987_030 |
0.1905277428 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 4 |
Hb_001006_280 |
0.2108931163 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |
| 5 |
Hb_020178_010 |
0.2243411425 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein A-like [Jatropha curcas] |
| 6 |
Hb_004965_060 |
0.2243761386 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: U4/U6 small nuclear ribonucleoprotein PRP4-like protein [Jatropha curcas] |
| 7 |
Hb_011114_030 |
0.2250786403 |
- |
- |
PREDICTED: probable carboxylesterase 15 [Jatropha curcas] |
| 8 |
Hb_000914_130 |
0.2292872552 |
- |
- |
copper-transporting atpase p-type, putative [Ricinus communis] |
| 9 |
Hb_000421_270 |
0.2321812196 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 1 [Theobroma cacao] |
| 10 |
Hb_002025_350 |
0.2322171266 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |
| 11 |
Hb_000116_590 |
0.2356292539 |
- |
- |
PREDICTED: splicing factor, suppressor of white-apricot homolog [Malus domestica] |
| 12 |
Hb_000556_180 |
0.2387423882 |
- |
- |
hypothetical protein POPTR_0014s06690g [Populus trichocarpa] |
| 13 |
Hb_000034_030 |
0.2391402766 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 14 |
Hb_008695_130 |
0.2391416894 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_008899_030 |
0.240806953 |
- |
- |
Lipase class 3-related protein, putative [Theobroma cacao] |
| 16 |
Hb_000130_080 |
0.2428997666 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_001024_030 |
0.2439206977 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000261_420 |
0.2447531693 |
- |
- |
PREDICTED: receptor like protein kinase S.2 [Jatropha curcas] |
| 19 |
Hb_008759_020 |
0.2456085235 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 20 |
Hb_025526_080 |
0.2461221268 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Vitis vinifera] |