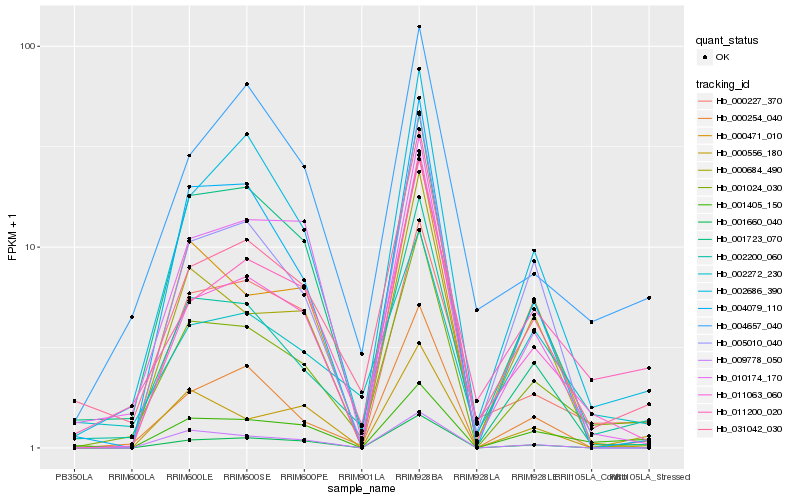
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001024_030 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_031042_030 |
0.1225716081 |
- |
- |
PREDICTED: WAT1-related protein At5g47470-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000254_040 |
0.1300331345 |
- |
- |
PREDICTED: uncharacterized protein LOC105634992 [Jatropha curcas] |
| 4 |
Hb_002686_390 |
0.1322419268 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 5 |
Hb_011200_020 |
0.1444522433 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 6 |
Hb_002200_060 |
0.1467359917 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8 [Jatropha curcas] |
| 7 |
Hb_011063_060 |
0.1490767692 |
- |
- |
PREDICTED: endochitinase-like [Jatropha curcas] |
| 8 |
Hb_001405_150 |
0.155444834 |
- |
- |
PREDICTED: protein At-4/1 [Jatropha curcas] |
| 9 |
Hb_004079_110 |
0.1569068645 |
- |
- |
PREDICTED: uncharacterized protein LOC100257802 [Vitis vinifera] |
| 10 |
Hb_001660_040 |
0.1631787462 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 11 |
Hb_000684_490 |
0.1644162555 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 44 [Jatropha curcas] |
| 12 |
Hb_004657_040 |
0.1650325545 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |
| 13 |
Hb_000471_010 |
0.1674292161 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_002272_230 |
0.169303834 |
- |
- |
sodium/hydrogen exchanger, putative [Ricinus communis] |
| 15 |
Hb_005010_040 |
0.169468873 |
- |
- |
PREDICTED: uncharacterized protein LOC105630824 [Jatropha curcas] |
| 16 |
Hb_000227_370 |
0.1712999976 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 17 |
Hb_001723_070 |
0.1713772196 |
- |
- |
PREDICTED: polygalacturonase At1g48100 [Jatropha curcas] |
| 18 |
Hb_000556_180 |
0.1718523398 |
- |
- |
hypothetical protein POPTR_0014s06690g [Populus trichocarpa] |
| 19 |
Hb_010174_170 |
0.1726114336 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X1 [Jatropha curcas] |
| 20 |
Hb_009778_050 |
0.1733418432 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |