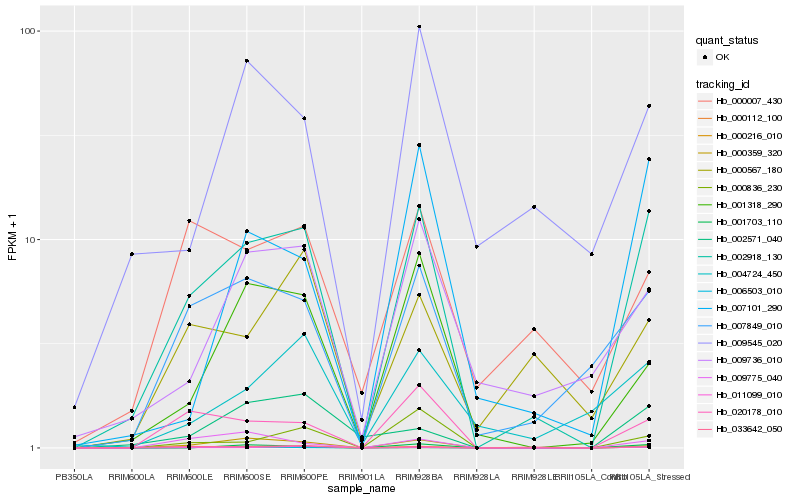
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002918_130 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 12 [Jatropha curcas] |
| 2 |
Hb_000112_100 |
0.209562647 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 3 |
Hb_007849_010 |
0.2145152667 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 4 |
Hb_020178_010 |
0.2267629797 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein A-like [Jatropha curcas] |
| 5 |
Hb_006503_010 |
0.2343880859 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 6 |
Hb_009736_010 |
0.2533351071 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_007101_290 |
0.257548376 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001703_110 |
0.2577702336 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 9 |
Hb_000216_010 |
0.2604881956 |
- |
- |
carbon monoxide dehydrogenase [Streptomyces sp. CNQ-509] |
| 10 |
Hb_000359_320 |
0.2605305296 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-4a-like [Jatropha curcas] |
| 11 |
Hb_011099_010 |
0.2639370206 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 12 |
Hb_001318_290 |
0.2641955148 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 13 |
Hb_000836_230 |
0.2663617457 |
- |
- |
Uncharacterized protein TCM_041879 [Theobroma cacao] |
| 14 |
Hb_033642_050 |
0.2690689481 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 4-like [Jatropha curcas] |
| 15 |
Hb_004724_450 |
0.2756102499 |
- |
- |
PREDICTED: uncharacterized protein LOC105647069 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000007_430 |
0.2772340111 |
- |
- |
hypothetical protein POPTR_0010s14280g [Populus trichocarpa] |
| 17 |
Hb_000567_180 |
0.2783976985 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.5 [Jatropha curcas] |
| 18 |
Hb_009775_040 |
0.2790654375 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Vitis vinifera] |
| 19 |
Hb_002571_040 |
0.280329465 |
- |
- |
hypothetical protein RCOM_1321060 [Ricinus communis] |
| 20 |
Hb_009545_020 |
0.2817398233 |
- |
- |
PREDICTED: uncharacterized protein LOC105632008 [Jatropha curcas] |