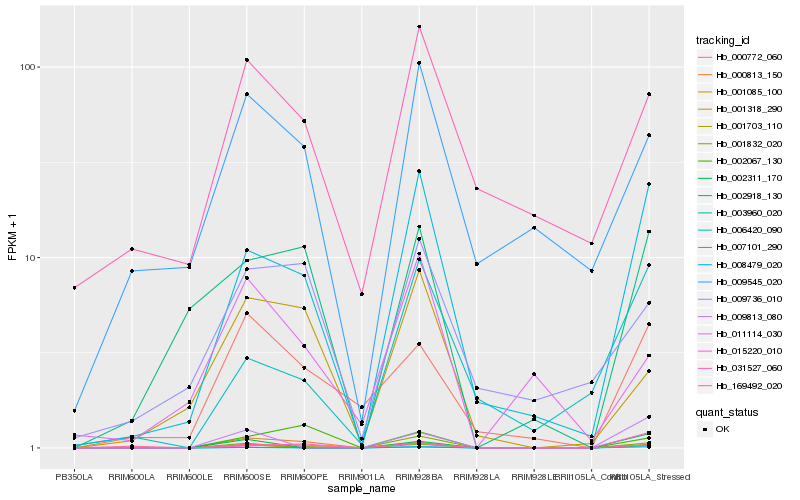
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001703_110 |
0.0 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 2 |
Hb_007101_290 |
0.1756992155 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000813_150 |
0.23301545 |
- |
- |
- |
| 4 |
Hb_003960_020 |
0.2423113977 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_169492_020 |
0.252126848 |
- |
- |
hypothetical protein JCGZ_11849 [Jatropha curcas] |
| 6 |
Hb_000772_060 |
0.2525903414 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002918_130 |
0.2577702336 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 12 [Jatropha curcas] |
| 8 |
Hb_002067_130 |
0.2623900628 |
- |
- |
putative non-LTR retroelement reverse transcriptase [Arabidopsis thaliana] |
| 9 |
Hb_001085_100 |
0.2676201034 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 10 |
Hb_001318_290 |
0.2788954448 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 11 |
Hb_008479_020 |
0.2885247602 |
- |
- |
PREDICTED: uncharacterized protein LOC105436302 [Cucumis sativus] |
| 12 |
Hb_001832_020 |
0.2994033191 |
- |
- |
PREDICTED: uncharacterized protein LOC104584029 [Brachypodium distachyon] |
| 13 |
Hb_009545_020 |
0.3032059875 |
- |
- |
PREDICTED: uncharacterized protein LOC105632008 [Jatropha curcas] |
| 14 |
Hb_009736_010 |
0.308694953 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_009813_080 |
0.3120169747 |
- |
- |
hypothetical protein CISIN_1g045991mg [Citrus sinensis] |
| 16 |
Hb_006420_090 |
0.312377819 |
- |
- |
PREDICTED: zinc finger protein 5 [Jatropha curcas] |
| 17 |
Hb_031527_060 |
0.3131147468 |
- |
- |
glutamate decarboxylase, putative [Ricinus communis] |
| 18 |
Hb_002311_170 |
0.3132077433 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 2-like [Eucalyptus grandis] |
| 19 |
Hb_015220_010 |
0.3191673594 |
- |
- |
PREDICTED: ribonuclease 3-like protein 3 [Jatropha curcas] |
| 20 |
Hb_011114_030 |
0.3197136281 |
- |
- |
PREDICTED: probable carboxylesterase 15 [Jatropha curcas] |