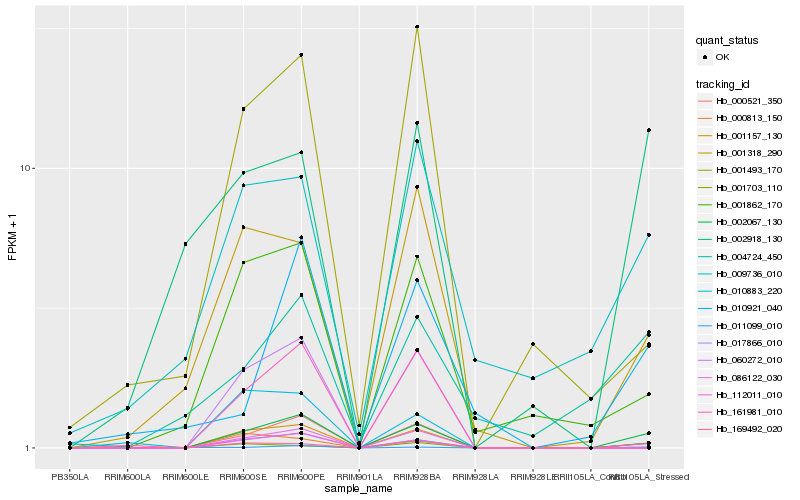
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002067_130 |
0.0 |
- |
- |
putative non-LTR retroelement reverse transcriptase [Arabidopsis thaliana] |
| 2 |
Hb_000813_150 |
0.2162209139 |
- |
- |
- |
| 3 |
Hb_001318_290 |
0.2560966437 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 4 |
Hb_001862_170 |
0.2589292648 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_011099_010 |
0.2617457432 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 6 |
Hb_001703_110 |
0.2623900628 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 7 |
Hb_002918_130 |
0.2839882961 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 12 [Jatropha curcas] |
| 8 |
Hb_010883_220 |
0.2902509555 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Prunus mume] |
| 9 |
Hb_009736_010 |
0.2904463736 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_010921_040 |
0.2916215278 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105634154 [Jatropha curcas] |
| 11 |
Hb_001493_170 |
0.2917714172 |
- |
- |
PREDICTED: uncharacterized protein LOC105631328 [Jatropha curcas] |
| 12 |
Hb_169492_020 |
0.2923046496 |
- |
- |
hypothetical protein JCGZ_11849 [Jatropha curcas] |
| 13 |
Hb_060272_010 |
0.2946085269 |
- |
- |
PREDICTED: gibberellin 20 oxidase 2 [Populus euphratica] |
| 14 |
Hb_086122_030 |
0.2949300019 |
- |
- |
D-glycerate 3-kinase, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_161981_010 |
0.2958798276 |
- |
- |
brassinosteroid insensitive 1 -like protein [Gossypium arboreum] |
| 16 |
Hb_004724_450 |
0.2969457933 |
- |
- |
PREDICTED: uncharacterized protein LOC105647069 isoform X1 [Jatropha curcas] |
| 17 |
Hb_017866_010 |
0.2970941566 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 18 |
Hb_001157_130 |
0.2996770079 |
- |
- |
PREDICTED: transmembrane protein 45B-like [Jatropha curcas] |
| 19 |
Hb_000521_350 |
0.2998546876 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_112011_010 |
0.3005291618 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |