| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
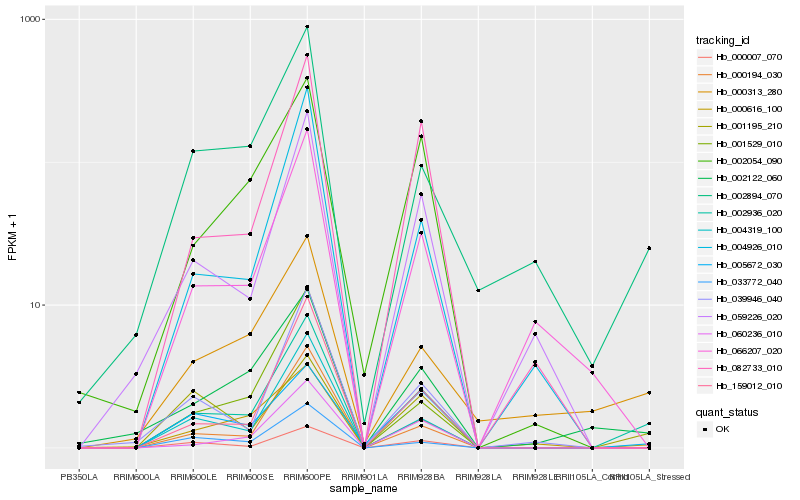
Hb_002936_020 |
0.0 |
- |
- |
hypothetical protein VITISV_001748 [Vitis vinifera] |
| 2 |
Hb_004319_100 |
0.1768082656 |
- |
- |
PREDICTED: gibberellin-regulated protein 14 isoform X3 [Jatropha curcas] |
| 3 |
Hb_002894_070 |
0.1792773621 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein B [Jatropha curcas] |
| 4 |
Hb_000616_100 |
0.1794043191 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 5 |
Hb_159012_010 |
0.1803486253 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g06940 [Jatropha curcas] |
| 6 |
Hb_000194_030 |
0.1859061878 |
transcription factor |
TF Family: NAC |
NAC transcription factor 059 [Jatropha curcas] |
| 7 |
Hb_001529_010 |
0.1897315759 |
- |
- |
- |
| 8 |
Hb_000313_280 |
0.1898216365 |
- |
- |
hypothetical protein PRUPE_ppa014856mg, partial [Prunus persica] |
| 9 |
Hb_060236_010 |
0.1911849171 |
- |
- |
hypothetical protein JCGZ_26099 [Jatropha curcas] |
| 10 |
Hb_002122_060 |
0.1922802952 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_000007_070 |
0.1926612559 |
- |
- |
hypothetical protein POPTR_0008s11290g [Populus trichocarpa] |
| 12 |
Hb_082733_010 |
0.1951025033 |
- |
- |
hypothetical protein JCGZ_23600 [Jatropha curcas] |
| 13 |
Hb_059226_020 |
0.1988070538 |
- |
- |
hypothetical protein B456_008G287700 [Gossypium raimondii] |
| 14 |
Hb_004926_010 |
0.1992859234 |
- |
- |
hypothetical protein JCGZ_27059 [Jatropha curcas] |
| 15 |
Hb_033772_040 |
0.1998525989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_039946_040 |
0.2030424774 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 17 |
Hb_066207_020 |
0.2059436512 |
- |
- |
hypothetical protein JCGZ_22048 [Jatropha curcas] |
| 18 |
Hb_005672_030 |
0.2077336279 |
- |
- |
PREDICTED: high mobility group B protein 3-like [Jatropha curcas] |
| 19 |
Hb_002054_090 |
0.208775593 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein-like [Gossypium raimondii] |
| 20 |
Hb_001195_210 |
0.2091411944 |
- |
- |
- |