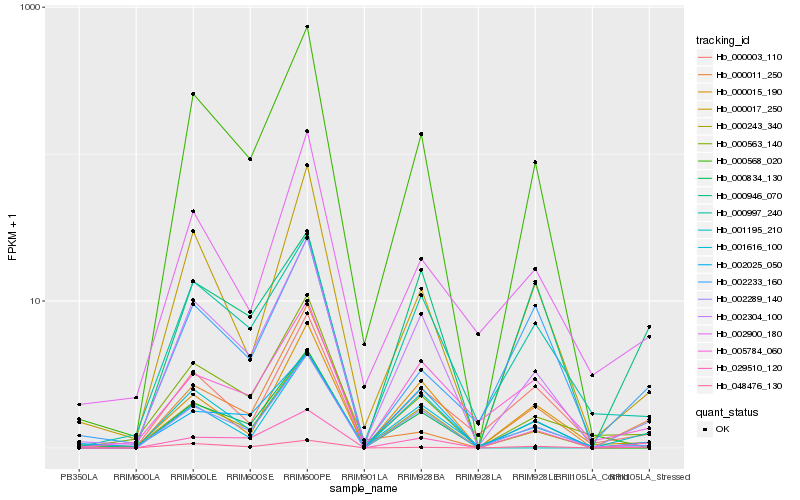
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002289_140 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000946_070 |
0.1853612222 |
- |
- |
chaperone protein DNAj, putative [Ricinus communis] |
| 3 |
Hb_029510_120 |
0.1897436679 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 8-like [Jatropha curcas] |
| 4 |
Hb_000003_110 |
0.1911171633 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 5 |
Hb_001616_100 |
0.1939592042 |
- |
- |
PREDICTED: cell division control protein 45 homolog [Jatropha curcas] |
| 6 |
Hb_000015_190 |
0.2002791464 |
- |
- |
PREDICTED: uncharacterized protein LOC105647810 [Jatropha curcas] |
| 7 |
Hb_000017_250 |
0.2031843821 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 8 |
Hb_048476_130 |
0.2034118211 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 9 |
Hb_005784_060 |
0.2049814347 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 10 |
Hb_000563_140 |
0.2067291607 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 11 |
Hb_000834_130 |
0.2085990763 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002025_050 |
0.2092777831 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002900_180 |
0.2126007114 |
transcription factor |
TF Family: LIM |
LIM1 [Hevea brasiliensis] |
| 14 |
Hb_002304_100 |
0.2131941394 |
- |
- |
hypothetical protein JCGZ_21524 [Jatropha curcas] |
| 15 |
Hb_000011_250 |
0.2141737212 |
- |
- |
PREDICTED: RING-H2 finger protein ATL3-like [Jatropha curcas] |
| 16 |
Hb_000243_340 |
0.2145639892 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 17 |
Hb_000568_020 |
0.2183578386 |
- |
- |
PREDICTED: probable aquaporin PIP2-5 [Jatropha curcas] |
| 18 |
Hb_000997_240 |
0.2186735986 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA4, putative [Ricinus communis] |
| 19 |
Hb_001195_210 |
0.2188822314 |
- |
- |
- |
| 20 |
Hb_002233_160 |
0.2189786642 |
- |
- |
PREDICTED: uncharacterized protein LOC105633094 [Jatropha curcas] |