| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
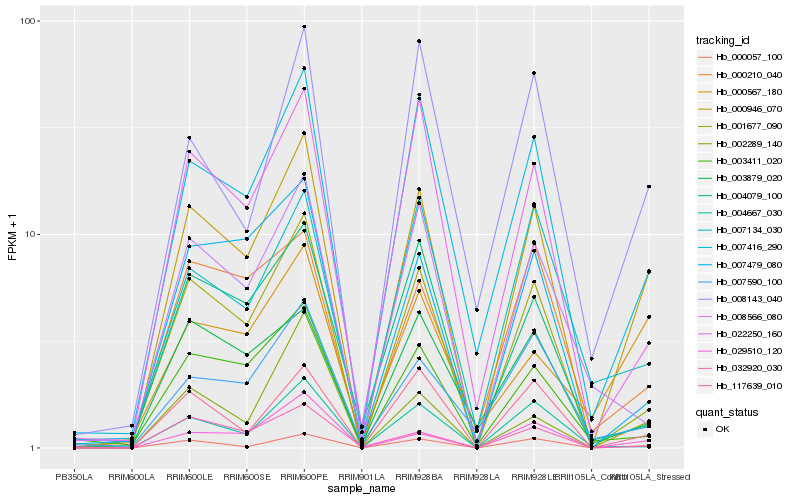
Hb_000946_070 |
0.0 |
- |
- |
chaperone protein DNAj, putative [Ricinus communis] |
| 2 |
Hb_007416_290 |
0.1297434809 |
- |
- |
PREDICTED: plasma membrane ATPase 4-like [Pyrus x bretschneideri] |
| 3 |
Hb_000057_100 |
0.1369934073 |
- |
- |
Peroxidase 57 precursor, putative [Ricinus communis] |
| 4 |
Hb_001677_090 |
0.1404403722 |
- |
- |
PREDICTED: CASP-like protein 5B2 [Jatropha curcas] |
| 5 |
Hb_029510_120 |
0.1523529586 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 8-like [Jatropha curcas] |
| 6 |
Hb_117639_010 |
0.1591060877 |
- |
- |
hypothetical protein B456_013G070100 [Gossypium raimondii] |
| 7 |
Hb_022250_160 |
0.1601708719 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |
| 8 |
Hb_007479_080 |
0.172900787 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003411_020 |
0.1729660523 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 10 |
Hb_008143_040 |
0.1779119459 |
- |
- |
Alpha-expansin 4 precursor, putative [Ricinus communis] |
| 11 |
Hb_000210_040 |
0.1817351669 |
- |
- |
PREDICTED: putative HVA22-like protein g [Jatropha curcas] |
| 12 |
Hb_002289_140 |
0.1853612222 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004079_100 |
0.1859677065 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 14 |
Hb_007590_100 |
0.187390403 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 15 |
Hb_000567_180 |
0.187460447 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.5 [Jatropha curcas] |
| 16 |
Hb_032920_030 |
0.1883641504 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 17 |
Hb_003879_020 |
0.1891157146 |
- |
- |
PREDICTED: MATE efflux family protein 1-like [Jatropha curcas] |
| 18 |
Hb_004667_030 |
0.1910673425 |
- |
- |
hypothetical protein VITISV_034568 [Vitis vinifera] |
| 19 |
Hb_008566_080 |
0.1912689145 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 20 |
Hb_007134_030 |
0.1923625021 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |