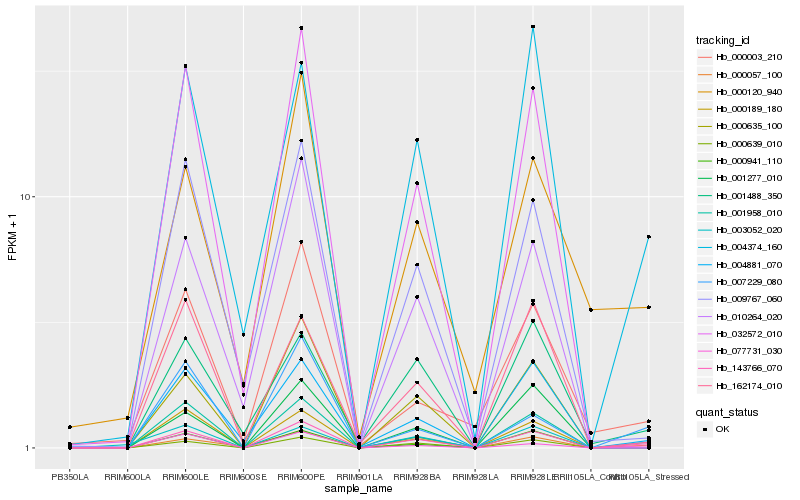
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_143766_070 |
0.0 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 2 |
Hb_000057_100 |
0.1623143018 |
- |
- |
Peroxidase 57 precursor, putative [Ricinus communis] |
| 3 |
Hb_004881_070 |
0.1633002978 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_004374_160 |
0.1663374369 |
transcription factor |
TF Family: bHLH |
hypothetical protein JCGZ_20587 [Jatropha curcas] |
| 5 |
Hb_003052_020 |
0.17217 |
- |
- |
PREDICTED: chaperonin 60 subunit beta 4, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_000635_100 |
0.1781709452 |
- |
- |
- |
| 7 |
Hb_000639_010 |
0.1790504172 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_09727 [Jatropha curcas] |
| 8 |
Hb_077731_030 |
0.1796942657 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 9 |
Hb_032572_010 |
0.1823921421 |
- |
- |
PREDICTED: phospholipase A2-alpha [Jatropha curcas] |
| 10 |
Hb_001488_350 |
0.1887470297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_009767_060 |
0.1919551919 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 12 |
Hb_007229_080 |
0.1940039056 |
- |
- |
Nonspecific lipid-transfer protein A, putative [Ricinus communis] |
| 13 |
Hb_001958_010 |
0.1941627024 |
- |
- |
hypothetical protein CICLE_v100078532mg, partial [Citrus clementina] |
| 14 |
Hb_162174_010 |
0.1982172515 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_000120_940 |
0.1999957949 |
- |
- |
PREDICTED: phosphoserine aminotransferase 1, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_010264_020 |
0.2001023603 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_000189_180 |
0.2012579312 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 18 |
Hb_000941_110 |
0.2020019126 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 19 |
Hb_000003_210 |
0.202607623 |
- |
- |
PREDICTED: GEM-like protein 4 [Jatropha curcas] |
| 20 |
Hb_001277_010 |
0.2029906462 |
- |
- |
unknown [Populus trichocarpa] |