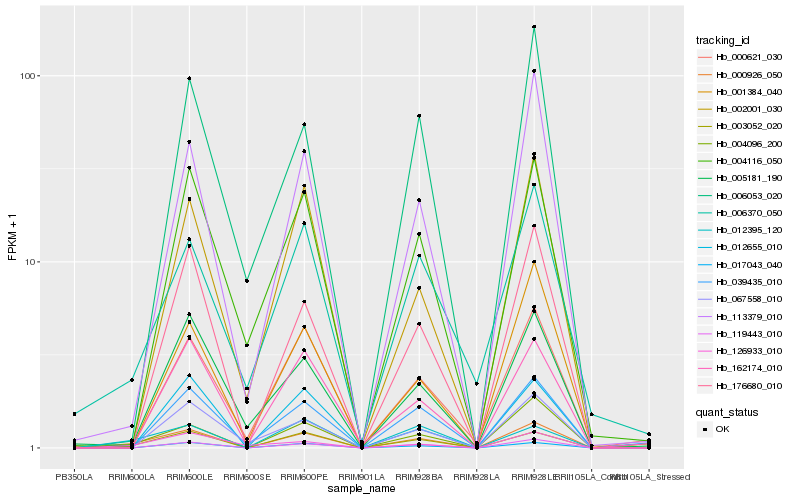
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000926_050 |
0.0 |
- |
- |
PREDICTED: vinorine synthase-like [Populus euphratica] |
| 2 |
Hb_004096_200 |
0.1206938926 |
- |
- |
PREDICTED: GDT1-like protein 5 [Eucalyptus grandis] |
| 3 |
Hb_119443_010 |
0.1703616219 |
- |
- |
hypothetical protein JCGZ_17348 [Jatropha curcas] |
| 4 |
Hb_012395_120 |
0.1798618231 |
- |
- |
- |
| 5 |
Hb_017043_040 |
0.1836267358 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 6 |
Hb_176680_010 |
0.1853227742 |
- |
- |
hypothetical protein JCGZ_22787 [Jatropha curcas] |
| 7 |
Hb_113379_010 |
0.1888233074 |
- |
- |
PREDICTED: beta-xylosidase/alpha-L-arabinofuranosidase 1 [Jatropha curcas] |
| 8 |
Hb_006370_050 |
0.1955760346 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_126933_010 |
0.1968094754 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 10 |
Hb_002001_030 |
0.2046647863 |
- |
- |
Plasma membrane ATPase 4 [Gossypium arboreum] |
| 11 |
Hb_039435_010 |
0.2047940699 |
- |
- |
hypothetical protein SORBIDRAFT_10g025530 [Sorghum bicolor] |
| 12 |
Hb_004116_050 |
0.2050170707 |
- |
- |
PREDICTED: cytochrome P450 90A1 [Jatropha curcas] |
| 13 |
Hb_162174_010 |
0.2050527 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_005181_190 |
0.2059191741 |
- |
- |
PREDICTED: uncharacterized protein LOC105643550 [Jatropha curcas] |
| 15 |
Hb_067558_010 |
0.2061938577 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 16 |
Hb_012655_010 |
0.2070734836 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 17 |
Hb_003052_020 |
0.2071117027 |
- |
- |
PREDICTED: chaperonin 60 subunit beta 4, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_001384_040 |
0.2078428032 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 19 |
Hb_000621_030 |
0.2088671002 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 20 |
Hb_006053_020 |
0.2093652024 |
- |
- |
CAP [Theobroma cacao] |