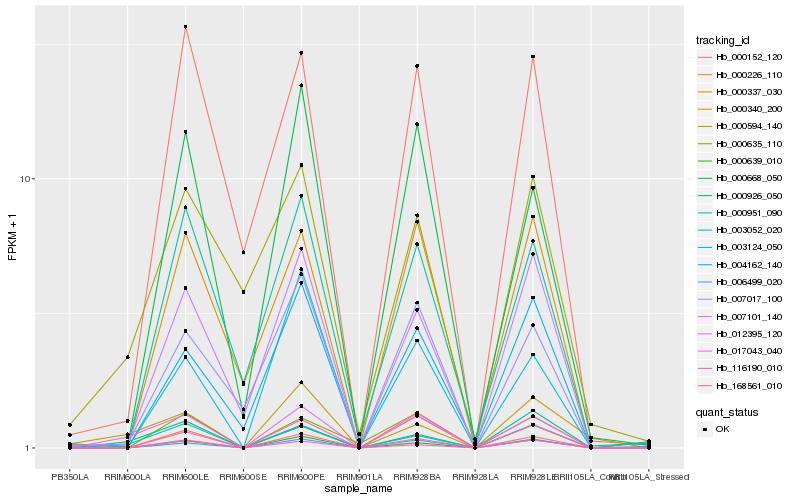
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012395_120 |
0.0 |
- |
- |
- |
| 2 |
Hb_000926_050 |
0.1798618231 |
- |
- |
PREDICTED: vinorine synthase-like [Populus euphratica] |
| 3 |
Hb_168561_010 |
0.1978753204 |
- |
- |
PREDICTED: notchless protein homolog [Jatropha curcas] |
| 4 |
Hb_006499_020 |
0.2007369373 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 5 |
Hb_000639_010 |
0.2034936843 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_09727 [Jatropha curcas] |
| 6 |
Hb_000668_050 |
0.2134602413 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 7 |
Hb_003052_020 |
0.2174312757 |
- |
- |
PREDICTED: chaperonin 60 subunit beta 4, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_007101_140 |
0.2178232697 |
- |
- |
hypothetical protein JCGZ_21777 [Jatropha curcas] |
| 9 |
Hb_000340_200 |
0.2222925237 |
- |
- |
PREDICTED: uncharacterized protein LOC105650039 [Jatropha curcas] |
| 10 |
Hb_116190_010 |
0.2236315498 |
- |
- |
PREDICTED: uncharacterized protein LOC105118535 [Populus euphratica] |
| 11 |
Hb_007017_100 |
0.2261385704 |
- |
- |
PREDICTED: uncharacterized protein LOC105649804 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000152_120 |
0.2276770148 |
- |
- |
alpha-l-fucosidase, putative [Ricinus communis] |
| 13 |
Hb_000594_140 |
0.2296112556 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 3 [Jatropha curcas] |
| 14 |
Hb_000951_090 |
0.2325997052 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 15 |
Hb_000226_110 |
0.2330675762 |
- |
- |
- |
| 16 |
Hb_017043_040 |
0.235798125 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 17 |
Hb_000337_030 |
0.2368771239 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 18 |
Hb_000635_110 |
0.2372667111 |
- |
- |
ELMO domain-containing protein, putative [Ricinus communis] |
| 19 |
Hb_003124_050 |
0.2381946127 |
- |
- |
PREDICTED: uncharacterized protein LOC105641435 [Jatropha curcas] |
| 20 |
Hb_004162_140 |
0.2389204255 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |