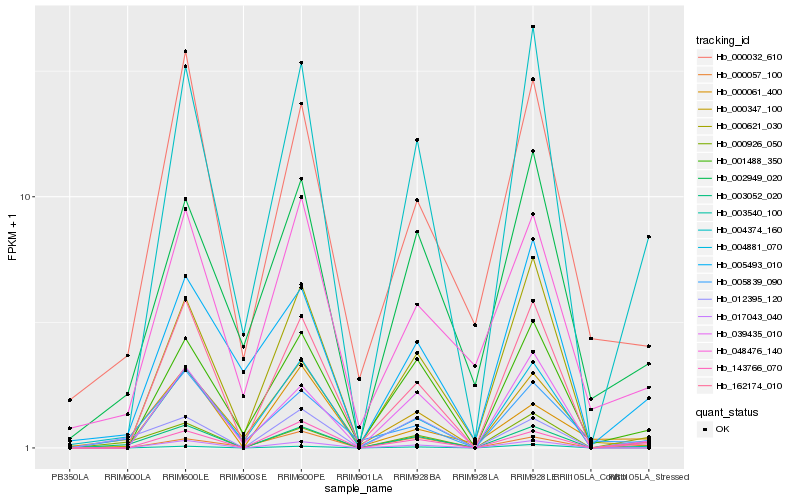
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003052_020 |
0.0 |
- |
- |
PREDICTED: chaperonin 60 subunit beta 4, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_004374_160 |
0.172062258 |
transcription factor |
TF Family: bHLH |
hypothetical protein JCGZ_20587 [Jatropha curcas] |
| 3 |
Hb_143766_070 |
0.17217 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 4 |
Hb_001488_350 |
0.2057357505 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_039435_010 |
0.2063758364 |
- |
- |
hypothetical protein SORBIDRAFT_10g025530 [Sorghum bicolor] |
| 6 |
Hb_000926_050 |
0.2071117027 |
- |
- |
PREDICTED: vinorine synthase-like [Populus euphratica] |
| 7 |
Hb_004881_070 |
0.2156877763 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_012395_120 |
0.2174312757 |
- |
- |
- |
| 9 |
Hb_162174_010 |
0.2183204299 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_000057_100 |
0.2211831248 |
- |
- |
Peroxidase 57 precursor, putative [Ricinus communis] |
| 11 |
Hb_002949_020 |
0.2219950042 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0002s23650g [Populus trichocarpa] |
| 12 |
Hb_005839_090 |
0.2222587664 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At3g56230 isoform X1 [Populus euphratica] |
| 13 |
Hb_017043_040 |
0.2291948 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 14 |
Hb_000621_030 |
0.2308361712 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005493_010 |
0.234793379 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_000032_610 |
0.2351480154 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase I, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_003540_100 |
0.236358296 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 18 |
Hb_048476_140 |
0.2378863303 |
- |
- |
PREDICTED: dynamin-related protein 1C [Jatropha curcas] |
| 19 |
Hb_000061_400 |
0.2381036458 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 20 |
Hb_000347_100 |
0.2392251694 |
transcription factor |
TF Family: C3H |
hypothetical protein RCOM_0684740 [Ricinus communis] |