| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
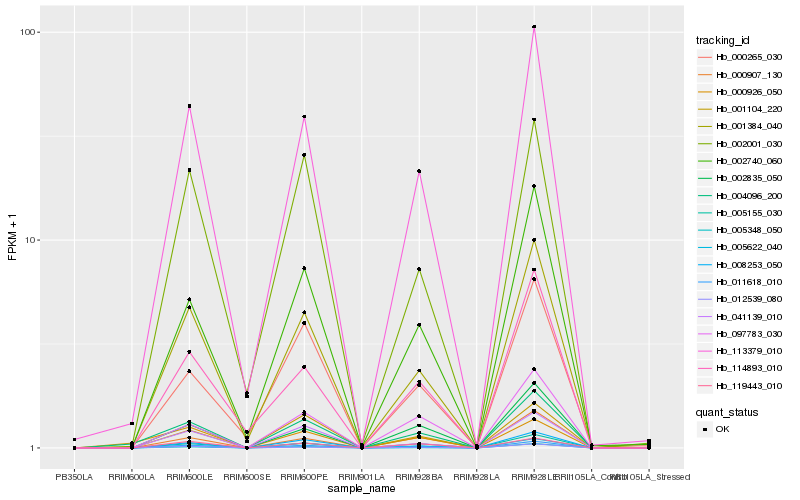
Hb_004096_200 |
0.0 |
- |
- |
PREDICTED: GDT1-like protein 5 [Eucalyptus grandis] |
| 2 |
Hb_113379_010 |
0.1111799997 |
- |
- |
PREDICTED: beta-xylosidase/alpha-L-arabinofuranosidase 1 [Jatropha curcas] |
| 3 |
Hb_000926_050 |
0.1206938926 |
- |
- |
PREDICTED: vinorine synthase-like [Populus euphratica] |
| 4 |
Hb_001384_040 |
0.1320301398 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 5 |
Hb_005155_030 |
0.1356875744 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 6 |
Hb_002740_060 |
0.1410551644 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 7 |
Hb_011618_010 |
0.1415540923 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 8 |
Hb_008253_050 |
0.1438914848 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0001s36220g [Populus trichocarpa] |
| 9 |
Hb_001104_220 |
0.1463145203 |
- |
- |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 10 |
Hb_005622_040 |
0.1468768691 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 11 |
Hb_041139_010 |
0.1500014528 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 12 |
Hb_012539_080 |
0.1561661772 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 13 |
Hb_097783_030 |
0.1569048638 |
- |
- |
PREDICTED: probable peroxidase 61 [Jatropha curcas] |
| 14 |
Hb_119443_010 |
0.1590508941 |
- |
- |
hypothetical protein JCGZ_17348 [Jatropha curcas] |
| 15 |
Hb_002835_050 |
0.1590836266 |
- |
- |
PREDICTED: high-affinity nitrate transporter 3.1-like [Jatropha curcas] |
| 16 |
Hb_002001_030 |
0.1627905991 |
- |
- |
Plasma membrane ATPase 4 [Gossypium arboreum] |
| 17 |
Hb_000907_130 |
0.1639593543 |
- |
- |
- |
| 18 |
Hb_000265_030 |
0.1684187233 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 19 |
Hb_005348_050 |
0.1707699831 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 20 |
Hb_114893_010 |
0.1708785645 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |