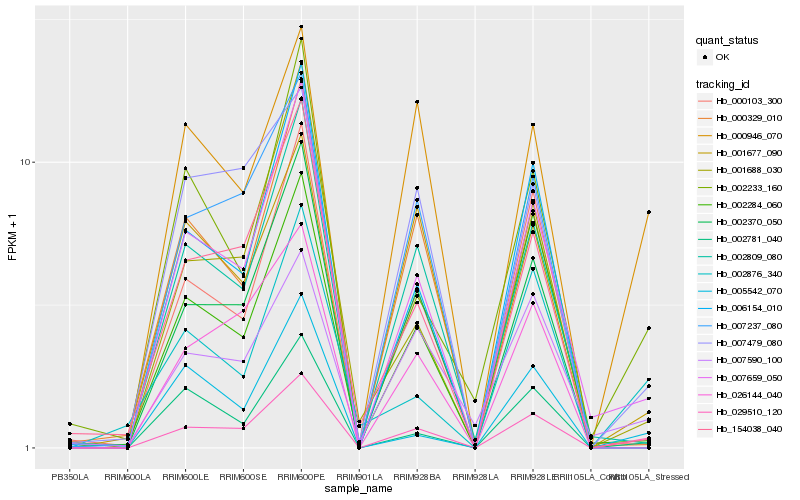
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029510_120 |
0.0 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 8-like [Jatropha curcas] |
| 2 |
Hb_007659_050 |
0.1141981322 |
- |
- |
PREDICTED: xylogen-like protein 11 [Jatropha curcas] |
| 3 |
Hb_000946_070 |
0.1523529586 |
- |
- |
chaperone protein DNAj, putative [Ricinus communis] |
| 4 |
Hb_002370_050 |
0.1591746762 |
- |
- |
hypothetical protein JCGZ_18849 [Jatropha curcas] |
| 5 |
Hb_002233_160 |
0.1610236619 |
- |
- |
PREDICTED: uncharacterized protein LOC105633094 [Jatropha curcas] |
| 6 |
Hb_001688_030 |
0.163848948 |
- |
- |
hypothetical protein JCGZ_10716 [Jatropha curcas] |
| 7 |
Hb_002809_080 |
0.1672348501 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002781_040 |
0.1700552426 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000103_300 |
0.1714631023 |
- |
- |
PREDICTED: perakine reductase-like [Jatropha curcas] |
| 10 |
Hb_006154_010 |
0.1724219058 |
- |
- |
UDP-glucose 6-dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_154038_040 |
0.1731258544 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 12 |
Hb_002284_060 |
0.1769364539 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 13 |
Hb_007237_080 |
0.177643935 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 14 |
Hb_002876_340 |
0.1776665742 |
- |
- |
hypothetical protein POPTR_0013s11770g [Populus trichocarpa] |
| 15 |
Hb_007590_100 |
0.1788319706 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 16 |
Hb_001677_090 |
0.1788367024 |
- |
- |
PREDICTED: CASP-like protein 5B2 [Jatropha curcas] |
| 17 |
Hb_005542_070 |
0.1789588517 |
- |
- |
hypothetical protein ZEAMMB73_851878 [Zea mays] |
| 18 |
Hb_026144_040 |
0.1790017884 |
- |
- |
- |
| 19 |
Hb_007479_080 |
0.179047071 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000329_010 |
0.1800008268 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |