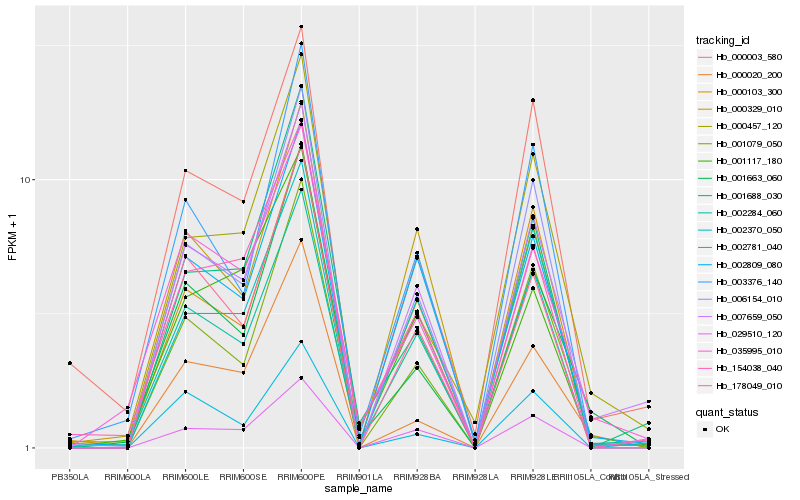
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007659_050 |
0.0 |
- |
- |
PREDICTED: xylogen-like protein 11 [Jatropha curcas] |
| 2 |
Hb_002809_080 |
0.1067866083 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001079_050 |
0.1106071357 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 4 |
Hb_029510_120 |
0.1141981322 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 8-like [Jatropha curcas] |
| 5 |
Hb_006154_010 |
0.119900594 |
- |
- |
UDP-glucose 6-dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_178049_010 |
0.1209543615 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_002370_050 |
0.1213561369 |
- |
- |
hypothetical protein JCGZ_18849 [Jatropha curcas] |
| 8 |
Hb_001688_030 |
0.125921986 |
- |
- |
hypothetical protein JCGZ_10716 [Jatropha curcas] |
| 9 |
Hb_000103_300 |
0.1264455996 |
- |
- |
PREDICTED: perakine reductase-like [Jatropha curcas] |
| 10 |
Hb_002781_040 |
0.1288981338 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000457_120 |
0.1305004354 |
- |
- |
PREDICTED: death-associated protein kinase dapk-1 [Jatropha curcas] |
| 12 |
Hb_001663_060 |
0.1326180153 |
- |
- |
ribonuclease t2, putative [Ricinus communis] |
| 13 |
Hb_035995_010 |
0.1330105847 |
- |
- |
- |
| 14 |
Hb_000329_010 |
0.1345554925 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 15 |
Hb_154038_040 |
0.1349730764 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 16 |
Hb_000020_200 |
0.1368685664 |
- |
- |
hypothetical protein EUGRSUZ_B00411 [Eucalyptus grandis] |
| 17 |
Hb_000003_580 |
0.1369714155 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002284_060 |
0.1382220863 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 19 |
Hb_001117_180 |
0.138756294 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003376_140 |
0.1395714434 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |