| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
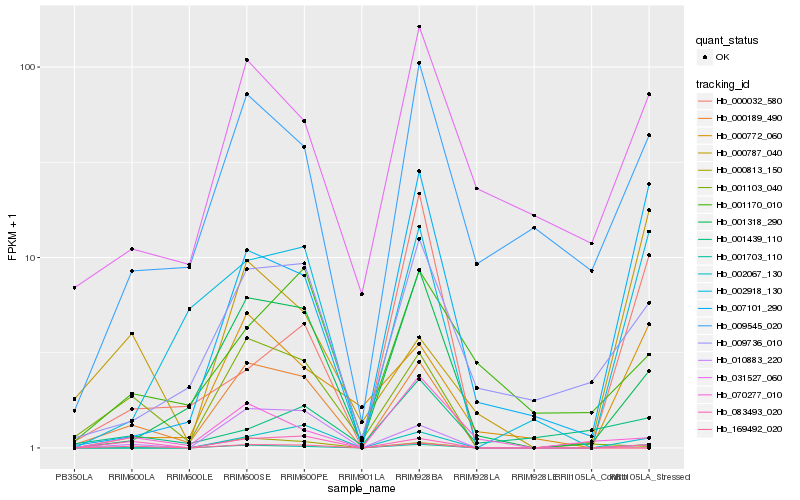
Hb_169492_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_11849 [Jatropha curcas] |
| 2 |
Hb_001703_110 |
0.252126848 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 3 |
Hb_002067_130 |
0.2923046496 |
- |
- |
putative non-LTR retroelement reverse transcriptase [Arabidopsis thaliana] |
| 4 |
Hb_001318_290 |
0.2977093292 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000813_150 |
0.2985054142 |
- |
- |
- |
| 6 |
Hb_002918_130 |
0.300093159 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 12 [Jatropha curcas] |
| 7 |
Hb_009545_020 |
0.3008645891 |
- |
- |
PREDICTED: uncharacterized protein LOC105632008 [Jatropha curcas] |
| 8 |
Hb_007101_290 |
0.3016039398 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_009736_010 |
0.3137787623 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000772_060 |
0.3171969859 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_031527_060 |
0.3222776825 |
- |
- |
glutamate decarboxylase, putative [Ricinus communis] |
| 12 |
Hb_070277_010 |
0.3343656558 |
- |
- |
PREDICTED: purine permease 3-like [Jatropha curcas] |
| 13 |
Hb_010883_220 |
0.3423859841 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Prunus mume] |
| 14 |
Hb_083493_020 |
0.3448115233 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Vitis vinifera] |
| 15 |
Hb_000032_580 |
0.3494766006 |
- |
- |
hypothetical protein RCOM_1311780 [Ricinus communis] |
| 16 |
Hb_001170_010 |
0.3495086575 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_001103_040 |
0.3524811539 |
- |
- |
hypothetical protein POPTR_0013s11170g [Populus trichocarpa] |
| 18 |
Hb_000189_490 |
0.3530899971 |
- |
- |
Sulfate/thiosulfate import ATP-binding protein cysA [Theobroma cacao] |
| 19 |
Hb_000787_040 |
0.3536692325 |
- |
- |
PREDICTED: uncharacterized protein LOC105115900 isoform X1 [Populus euphratica] |
| 20 |
Hb_001439_110 |
0.3541577861 |
transcription factor |
TF Family: ERF |
PREDICTED: uncharacterized protein LOC105133481 [Populus euphratica] |