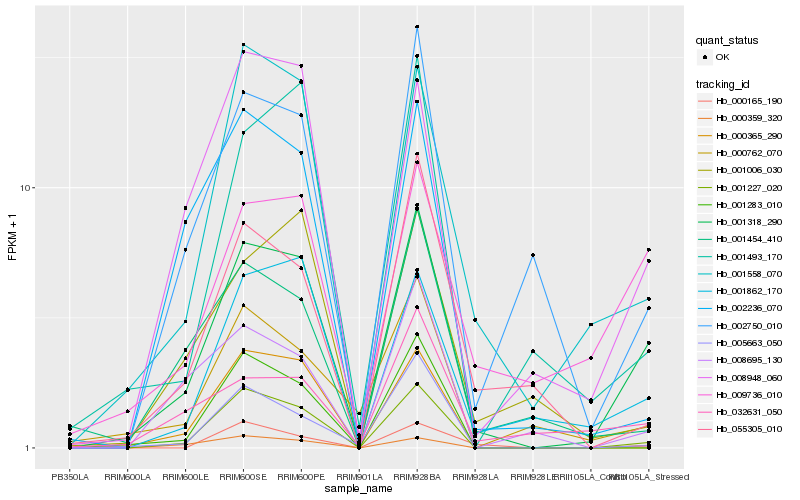
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001318_290 |
0.0 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 2 |
Hb_001227_020 |
0.1574834646 |
- |
- |
- |
| 3 |
Hb_000359_320 |
0.1720629727 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-4a-like [Jatropha curcas] |
| 4 |
Hb_001558_070 |
0.1758884976 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 5 |
Hb_008948_060 |
0.1857628854 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001862_170 |
0.193839342 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_009736_010 |
0.1947503947 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002750_010 |
0.2018799656 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |
| 9 |
Hb_001493_170 |
0.2088533713 |
- |
- |
PREDICTED: uncharacterized protein LOC105631328 [Jatropha curcas] |
| 10 |
Hb_032631_050 |
0.2182478981 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 11 |
Hb_005663_050 |
0.2248098857 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_001006_030 |
0.2276618538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000365_290 |
0.2284305064 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001454_410 |
0.2293294997 |
- |
- |
PREDICTED: uncharacterized protein LOC105643453 [Jatropha curcas] |
| 15 |
Hb_001283_010 |
0.2304451469 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 15 [Populus tremula x Populus tremuloides] |
| 16 |
Hb_002236_070 |
0.2308685244 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH36-like [Jatropha curcas] |
| 17 |
Hb_008695_130 |
0.2312879707 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_055305_010 |
0.2320646398 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 19 |
Hb_000762_070 |
0.2324320117 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 20 |
Hb_000165_190 |
0.2378702702 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |