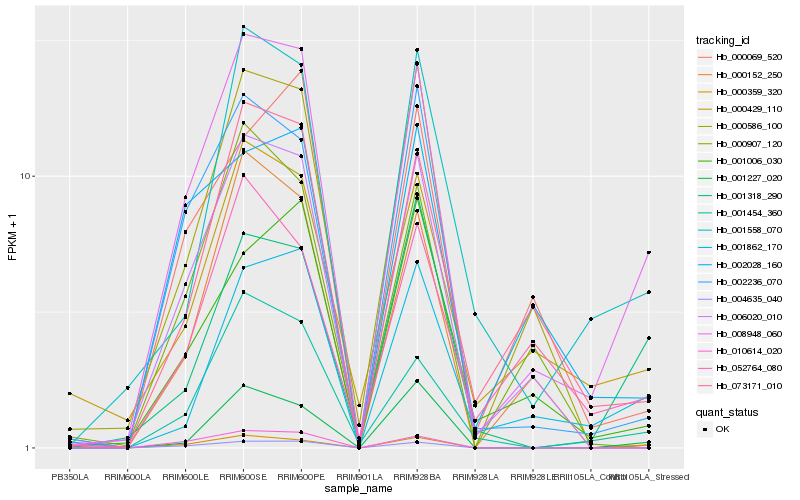
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008948_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000359_320 |
0.1328662114 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-4a-like [Jatropha curcas] |
| 3 |
Hb_001454_360 |
0.1573116413 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET17 [Jatropha curcas] |
| 4 |
Hb_073171_010 |
0.1592122642 |
- |
- |
PREDICTED: ABC transporter G family member 29-like [Jatropha curcas] |
| 5 |
Hb_002236_070 |
0.1639794254 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH36-like [Jatropha curcas] |
| 6 |
Hb_006020_010 |
0.1640906659 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 7 |
Hb_001862_170 |
0.1656212601 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_001558_070 |
0.1702186723 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 9 |
Hb_000429_110 |
0.1742207626 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein WIP2-like [Populus euphratica] |
| 10 |
Hb_004635_040 |
0.179931355 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 11 |
Hb_000152_250 |
0.1842840791 |
- |
- |
PREDICTED: uncharacterized protein LOC105648569 [Jatropha curcas] |
| 12 |
Hb_002028_160 |
0.1843426775 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL4-like [Populus euphratica] |
| 13 |
Hb_001318_290 |
0.1857628854 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 14 |
Hb_052764_080 |
0.1861588384 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD5 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001006_030 |
0.1861817774 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_010614_020 |
0.1870058467 |
- |
- |
BnaC05g27570D [Brassica napus] |
| 17 |
Hb_000069_520 |
0.1875578937 |
- |
- |
PREDICTED: transcription factor TRY-like [Jatropha curcas] |
| 18 |
Hb_001227_020 |
0.188902028 |
- |
- |
- |
| 19 |
Hb_000907_120 |
0.1910138846 |
transcription factor |
TF Family: G2-like |
PREDICTED: transcription repressor KAN1-like [Populus euphratica] |
| 20 |
Hb_000586_100 |
0.1920423515 |
- |
- |
conserved hypothetical protein [Ricinus communis] |