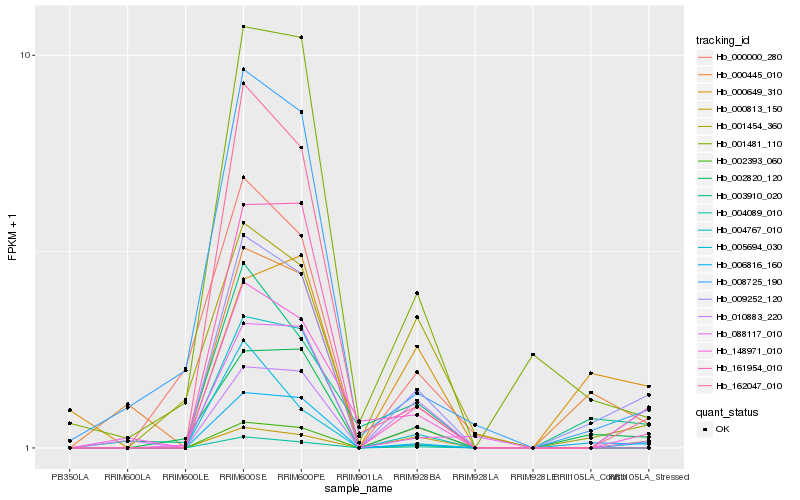
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009252_120 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_006816_160 |
0.1455902352 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 3 |
Hb_002820_120 |
0.1589152572 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 4 |
Hb_003910_020 |
0.1993272835 |
- |
- |
PREDICTED: reticulon-like protein B13 [Jatropha curcas] |
| 5 |
Hb_088117_010 |
0.2210683585 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 6 |
Hb_162047_010 |
0.221832344 |
- |
- |
PREDICTED: protein indeterminate-domain 16 [Jatropha curcas] |
| 7 |
Hb_000813_150 |
0.2288387545 |
- |
- |
- |
| 8 |
Hb_000445_010 |
0.2315940377 |
- |
- |
PREDICTED: uncharacterized protein LOC105113411 [Populus euphratica] |
| 9 |
Hb_001481_110 |
0.235963807 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 10 |
Hb_002393_060 |
0.2360585389 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 11 |
Hb_000649_310 |
0.2391200596 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 12 |
Hb_004089_010 |
0.239621844 |
- |
- |
hypothetical protein MIMGU_mgv1a018902mg [Erythranthe guttata] |
| 13 |
Hb_148971_010 |
0.2406680277 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 14 |
Hb_010883_220 |
0.2471548405 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Prunus mume] |
| 15 |
Hb_004767_010 |
0.247572714 |
- |
- |
hypothetical protein CICLE_v10010736mg [Citrus clementina] |
| 16 |
Hb_161954_010 |
0.2510774791 |
- |
- |
hypothetical protein JCGZ_05404 [Jatropha curcas] |
| 17 |
Hb_000000_280 |
0.251212575 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001454_360 |
0.2542848638 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET17 [Jatropha curcas] |
| 19 |
Hb_008725_190 |
0.2567332861 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |
| 20 |
Hb_005694_030 |
0.2572784038 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase PUB22-like [Jatropha curcas] |