| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
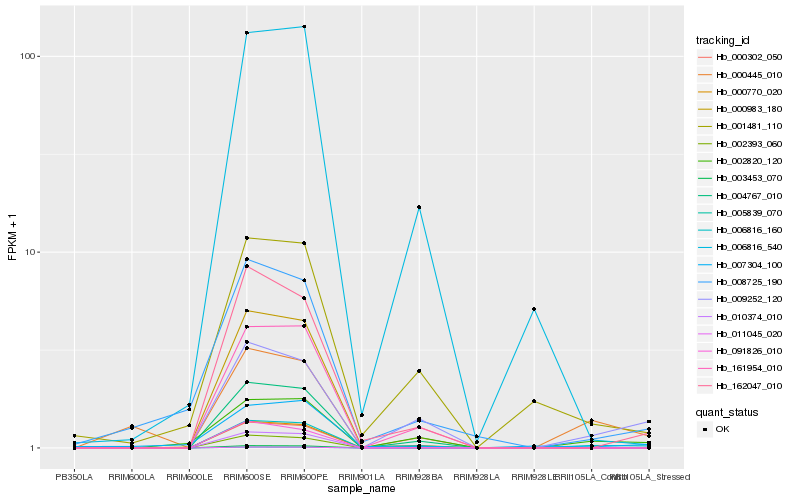
Hb_006816_160 |
0.0 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 2 |
Hb_009252_120 |
0.1455902352 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002820_120 |
0.1612611315 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 4 |
Hb_162047_010 |
0.1700910042 |
- |
- |
PREDICTED: protein indeterminate-domain 16 [Jatropha curcas] |
| 5 |
Hb_004767_010 |
0.1880161764 |
- |
- |
hypothetical protein CICLE_v10010736mg [Citrus clementina] |
| 6 |
Hb_161954_010 |
0.1913085003 |
- |
- |
hypothetical protein JCGZ_05404 [Jatropha curcas] |
| 7 |
Hb_000445_010 |
0.1926169751 |
- |
- |
PREDICTED: uncharacterized protein LOC105113411 [Populus euphratica] |
| 8 |
Hb_002393_060 |
0.1967090991 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 9 |
Hb_091826_010 |
0.2048020572 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000983_180 |
0.2057801548 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_001481_110 |
0.2115112635 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 12 |
Hb_008725_190 |
0.2134588692 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |
| 13 |
Hb_007304_100 |
0.2143410421 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Jatropha curcas] |
| 14 |
Hb_003453_070 |
0.2171013404 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor LEP [Jatropha curcas] |
| 15 |
Hb_005839_070 |
0.2171110786 |
- |
- |
hypothetical protein POPTR_0013s08160g [Populus trichocarpa] |
| 16 |
Hb_010374_010 |
0.2171172571 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 17 |
Hb_000770_020 |
0.217343018 |
- |
- |
hypothetical protein POPTR_0001s39195g [Populus trichocarpa] |
| 18 |
Hb_011045_020 |
0.217398018 |
- |
- |
hypothetical protein CICLE_v100140161mg, partial [Citrus clementina] |
| 19 |
Hb_006816_540 |
0.2174434009 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET16-like [Jatropha curcas] |
| 20 |
Hb_000302_050 |
0.2174594035 |
- |
- |
- |