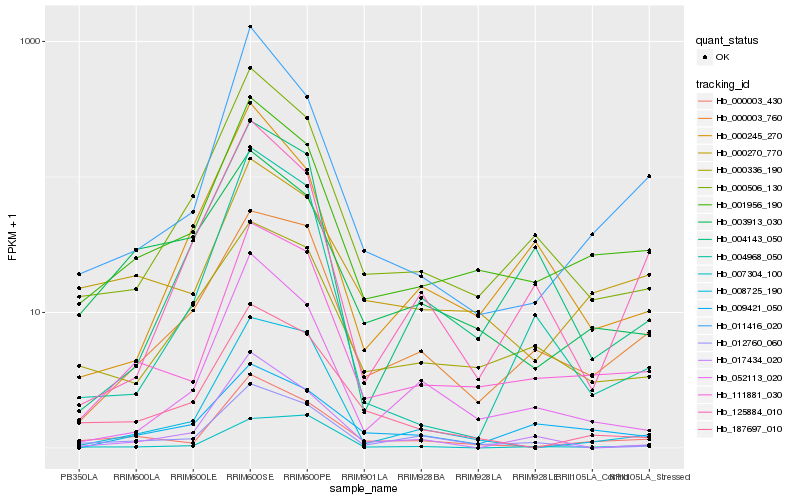
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_111881_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645255 [Jatropha curcas] |
| 2 |
Hb_001956_190 |
0.0965601073 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 3 |
Hb_000003_430 |
0.1230045937 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 4 |
Hb_000003_760 |
0.1352287612 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA26-like [Jatropha curcas] |
| 5 |
Hb_000506_130 |
0.1517504682 |
- |
- |
DGCR14-related family protein [Populus trichocarpa] |
| 6 |
Hb_052113_020 |
0.1678029205 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 7 |
Hb_008725_190 |
0.1749862156 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |
| 8 |
Hb_012760_060 |
0.1754669015 |
- |
- |
PREDICTED: uncharacterized protein LOC105647085 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000270_770 |
0.1771555995 |
- |
- |
PREDICTED: uncharacterized protein LOC105640410 [Jatropha curcas] |
| 10 |
Hb_000336_190 |
0.1786455388 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Gossypium raimondii] |
| 11 |
Hb_011416_020 |
0.1797674861 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_009421_050 |
0.1835816644 |
- |
- |
PREDICTED: uncharacterized protein LOC105647037 [Jatropha curcas] |
| 13 |
Hb_004143_050 |
0.1853892435 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 14 |
Hb_187697_010 |
0.1910140749 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 15 |
Hb_007304_100 |
0.1963692681 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Jatropha curcas] |
| 16 |
Hb_017434_020 |
0.201619778 |
transcription factor |
TF Family: C2H2 |
Zinc finger protein 4 [Populus trichocarpa] |
| 17 |
Hb_000245_270 |
0.2017347131 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 18 |
Hb_004968_050 |
0.202412875 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 19 |
Hb_125884_010 |
0.2056831369 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Jatropha curcas] |
| 20 |
Hb_003913_030 |
0.2058333974 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |