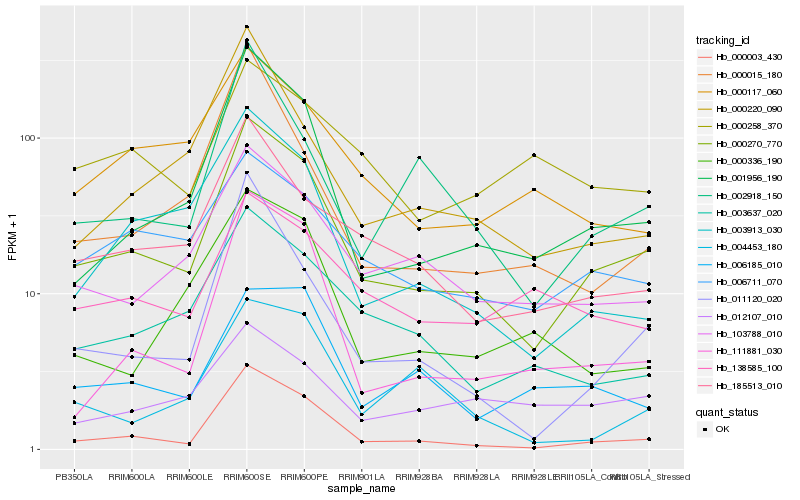
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_770 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640410 [Jatropha curcas] |
| 2 |
Hb_000003_430 |
0.1305163243 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 3 |
Hb_001956_190 |
0.1365252283 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 4 |
Hb_103788_010 |
0.1495519234 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 5 |
Hb_185513_010 |
0.1535669718 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_006711_070 |
0.1612236168 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 7 |
Hb_003637_020 |
0.1620836537 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003913_030 |
0.166010186 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 9 |
Hb_000117_060 |
0.1739469194 |
- |
- |
PREDICTED: uncharacterized protein LOC103949199 [Pyrus x bretschneideri] |
| 10 |
Hb_111881_030 |
0.1771555995 |
- |
- |
PREDICTED: uncharacterized protein LOC105645255 [Jatropha curcas] |
| 11 |
Hb_002918_150 |
0.1799405644 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 1 [Hevea brasiliensis] |
| 12 |
Hb_011120_020 |
0.1804701297 |
- |
- |
PREDICTED: casein kinase I-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_138585_100 |
0.1822839915 |
- |
- |
PREDICTED: uncharacterized protein LOC105648740 [Jatropha curcas] |
| 14 |
Hb_006185_010 |
0.1849002427 |
- |
- |
hypothetical protein L484_013453 [Morus notabilis] |
| 15 |
Hb_000220_090 |
0.1856975649 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Jatropha curcas] |
| 16 |
Hb_012107_010 |
0.1876257508 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 17 |
Hb_000336_190 |
0.1897732688 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Gossypium raimondii] |
| 18 |
Hb_000258_370 |
0.1990106054 |
- |
- |
hypothetical protein [Ricinus communis] |
| 19 |
Hb_000015_180 |
0.2003905817 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 20 |
Hb_004453_180 |
0.2016032697 |
- |
- |
hypothetical protein JCGZ_07188 [Jatropha curcas] |