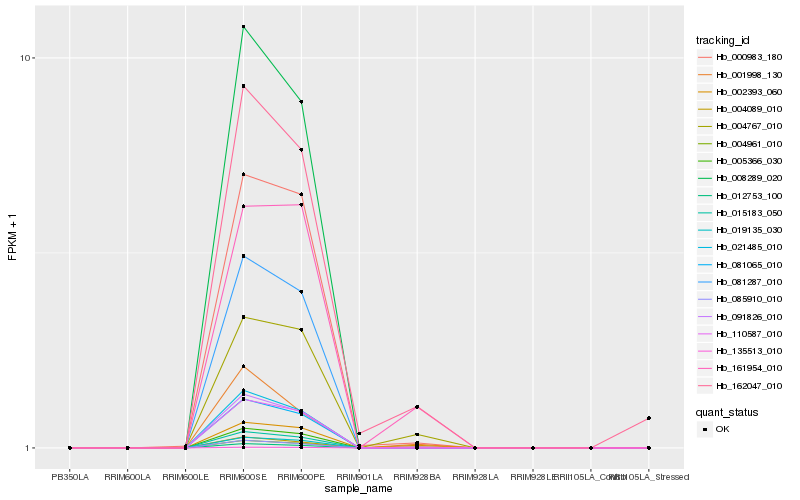
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_162047_010 |
0.0 |
- |
- |
PREDICTED: protein indeterminate-domain 16 [Jatropha curcas] |
| 2 |
Hb_091826_010 |
0.1130722148 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_004767_010 |
0.1268748505 |
- |
- |
hypothetical protein CICLE_v10010736mg [Citrus clementina] |
| 4 |
Hb_002393_060 |
0.1298245018 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 5 |
Hb_000983_180 |
0.1336822803 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 6 |
Hb_004089_010 |
0.1395611532 |
- |
- |
hypothetical protein MIMGU_mgv1a018902mg [Erythranthe guttata] |
| 7 |
Hb_161954_010 |
0.1476190482 |
- |
- |
hypothetical protein JCGZ_05404 [Jatropha curcas] |
| 8 |
Hb_001998_130 |
0.1481374586 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 35 family protein [Populus trichocarpa] |
| 9 |
Hb_015183_050 |
0.1484421396 |
- |
- |
hypothetical protein JCGZ_24010 [Jatropha curcas] |
| 10 |
Hb_081065_010 |
0.1485402556 |
- |
- |
hypothetical protein PHAVU_001G084100g, partial [Phaseolus vulgaris] |
| 11 |
Hb_004961_010 |
0.1488066383 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 12 |
Hb_008289_020 |
0.1488772978 |
- |
- |
PREDICTED: uncharacterized protein LOC105119066 [Populus euphratica] |
| 13 |
Hb_021485_010 |
0.1489185961 |
- |
- |
Integrase-type DNA-binding superfamily protein, putative [Theobroma cacao] |
| 14 |
Hb_005366_030 |
0.1496925525 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 15 |
Hb_019135_030 |
0.149909648 |
- |
- |
putative mitogen-activated protein kinase 1 [Populus trichocarpa] |
| 16 |
Hb_012753_100 |
0.1501053763 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_081287_010 |
0.1501281768 |
- |
- |
PREDICTED: alpha-dioxygenase 2 [Jatropha curcas] |
| 18 |
Hb_110587_010 |
0.1506079566 |
- |
- |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_135513_010 |
0.1512331864 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_085910_010 |
0.1522916871 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |