| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_081065_010 |
0.0 |
- |
- |
hypothetical protein PHAVU_001G084100g, partial [Phaseolus vulgaris] |
| 2 |
Hb_015183_050 |
0.0061726107 |
- |
- |
hypothetical protein JCGZ_24010 [Jatropha curcas] |
| 3 |
Hb_005366_030 |
0.0140380498 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 4 |
Hb_019135_030 |
0.015672272 |
- |
- |
putative mitogen-activated protein kinase 1 [Populus trichocarpa] |
| 5 |
Hb_004961_010 |
0.0160473721 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
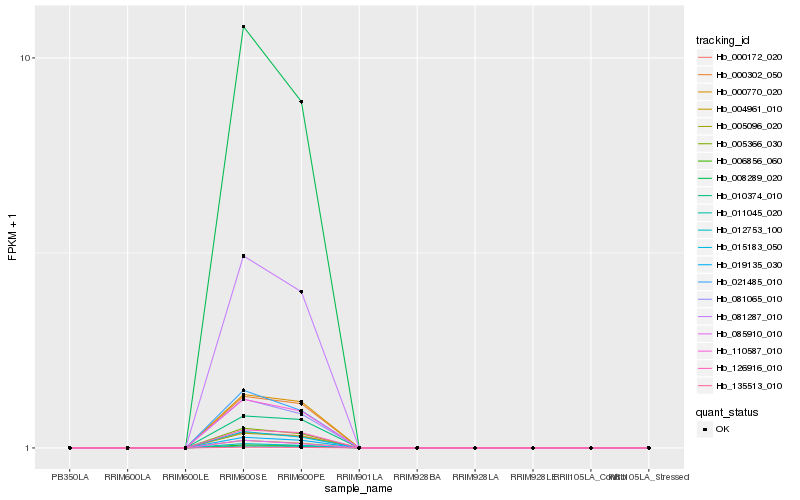
| 6 |
Hb_008289_020 |
0.0170219362 |
- |
- |
PREDICTED: uncharacterized protein LOC105119066 [Populus euphratica] |
| 7 |
Hb_081287_010 |
0.0172009368 |
- |
- |
PREDICTED: alpha-dioxygenase 2 [Jatropha curcas] |
| 8 |
Hb_021485_010 |
0.0175553041 |
- |
- |
Integrase-type DNA-binding superfamily protein, putative [Theobroma cacao] |
| 9 |
Hb_110587_010 |
0.0202473345 |
- |
- |
unnamed protein product [Coffea canephora] |
| 10 |
Hb_135513_010 |
0.0237570513 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_012753_100 |
0.0280424799 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000172_020 |
0.0338687672 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 3, mitochondrial [Vitis vinifera] |
| 13 |
Hb_005096_020 |
0.0366828194 |
- |
- |
PREDICTED: DIS3-like exonuclease 2-like isoform X1 [Citrus sinensis] |
| 14 |
Hb_085910_010 |
0.0399094662 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 15 |
Hb_006856_060 |
0.0458453347 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 16 |
Hb_126916_010 |
0.0490086536 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110, partial [Jatropha curcas] |
| 17 |
Hb_000302_050 |
0.0496287147 |
- |
- |
- |
| 18 |
Hb_011045_020 |
0.0507662184 |
- |
- |
hypothetical protein CICLE_v100140161mg, partial [Citrus clementina] |
| 19 |
Hb_000770_020 |
0.0518881246 |
- |
- |
hypothetical protein POPTR_0001s39195g [Populus trichocarpa] |
| 20 |
Hb_010374_010 |
0.0593752907 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |