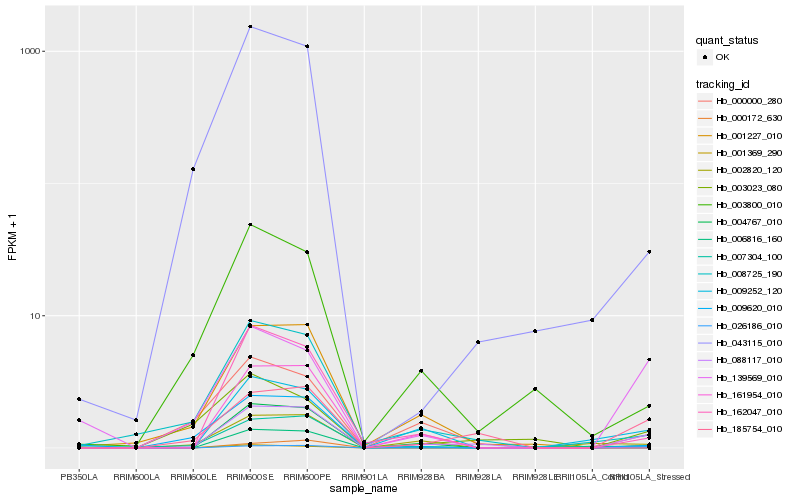
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_088117_010 |
0.0 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 2 |
Hb_185754_010 |
0.126420584 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 3 |
Hb_009252_120 |
0.2210683585 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000172_630 |
0.2225728102 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 5 |
Hb_026186_010 |
0.2249237657 |
- |
- |
hypothetical protein JCGZ_06344 [Jatropha curcas] |
| 6 |
Hb_006816_160 |
0.2311390892 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 7 |
Hb_008725_190 |
0.2382954371 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |
| 8 |
Hb_000000_280 |
0.238467016 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001369_290 |
0.239505724 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 10 |
Hb_162047_010 |
0.2399387251 |
- |
- |
PREDICTED: protein indeterminate-domain 16 [Jatropha curcas] |
| 11 |
Hb_002820_120 |
0.2526872556 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 12 |
Hb_007304_100 |
0.2560875585 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Jatropha curcas] |
| 13 |
Hb_139569_010 |
0.2576941643 |
- |
- |
hypothetical protein JCGZ_24635 [Jatropha curcas] |
| 14 |
Hb_003800_010 |
0.2679935572 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 15 |
Hb_003023_080 |
0.2686105791 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_043115_010 |
0.2723679741 |
- |
- |
hypothetical protein JCGZ_16710 [Jatropha curcas] |
| 17 |
Hb_009620_010 |
0.2725575673 |
transcription factor |
TF Family: Orphans |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004767_010 |
0.2726134143 |
- |
- |
hypothetical protein CICLE_v10010736mg [Citrus clementina] |
| 19 |
Hb_161954_010 |
0.2727097602 |
- |
- |
hypothetical protein JCGZ_05404 [Jatropha curcas] |
| 20 |
Hb_001227_010 |
0.2759737874 |
transcription factor |
TF Family: TCP |
hypothetical protein POPTR_0015s03140g [Populus trichocarpa] |