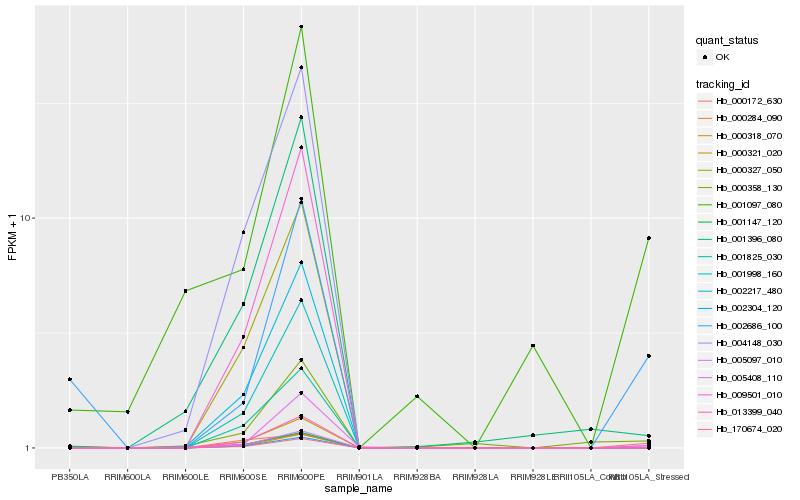
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005097_010 |
0.0 |
- |
- |
PREDICTED: aluminum-activated malate transporter 7-like isoform X1 [Populus euphratica] |
| 2 |
Hb_005408_110 |
0.217560014 |
- |
- |
purine permease, putative [Ricinus communis] |
| 3 |
Hb_002686_100 |
0.2216617658 |
- |
- |
- |
| 4 |
Hb_001097_080 |
0.2239118476 |
transcription factor |
TF Family: MYB-related |
triptychon and cpc, putative [Ricinus communis] |
| 5 |
Hb_000172_630 |
0.2266274999 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 6 |
Hb_000358_130 |
0.2318935374 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 7 |
Hb_013399_040 |
0.2350045517 |
- |
- |
hypothetical protein JCGZ_24713 [Jatropha curcas] |
| 8 |
Hb_170674_020 |
0.2350622599 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 9 |
Hb_000327_050 |
0.2351625801 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.7-like isoform X1 [Populus euphratica] |
| 10 |
Hb_000318_070 |
0.2352690994 |
- |
- |
PREDICTED: peroxidase 10-like [Jatropha curcas] |
| 11 |
Hb_002304_120 |
0.2355442444 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000321_020 |
0.2360265793 |
- |
- |
PREDICTED: uncharacterized protein LOC104891638 [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_001998_160 |
0.2365825592 |
- |
- |
PREDICTED: origin recognition complex subunit 1-like [Populus euphratica] |
| 14 |
Hb_002217_480 |
0.2380452238 |
- |
- |
PREDICTED: uncharacterized protein LOC105115446 [Populus euphratica] |
| 15 |
Hb_001147_120 |
0.2386735238 |
- |
- |
hypothetical protein JCGZ_14802 [Jatropha curcas] |
| 16 |
Hb_001825_030 |
0.2393763627 |
transcription factor |
TF Family: C2H2 |
PREDICTED: transcriptional regulator SUPERMAN [Jatropha curcas] |
| 17 |
Hb_004148_030 |
0.239383584 |
- |
- |
PREDICTED: protein RAFTIN 1B-like [Gossypium raimondii] |
| 18 |
Hb_000284_090 |
0.2394954674 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 19 |
Hb_009501_010 |
0.2396896226 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 20 |
Hb_001396_080 |
0.2416221244 |
- |
- |
conserved hypothetical protein [Ricinus communis] |