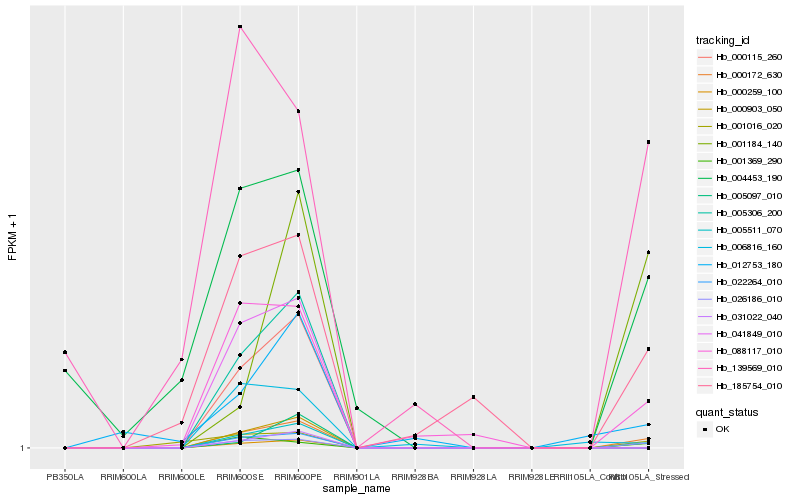
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000172_630 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 2 |
Hb_026186_010 |
0.1039875792 |
- |
- |
hypothetical protein JCGZ_06344 [Jatropha curcas] |
| 3 |
Hb_088117_010 |
0.2225728102 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 4 |
Hb_005097_010 |
0.2266274999 |
- |
- |
PREDICTED: aluminum-activated malate transporter 7-like isoform X1 [Populus euphratica] |
| 5 |
Hb_001369_290 |
0.2394938435 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 6 |
Hb_185754_010 |
0.2474251069 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 7 |
Hb_001016_020 |
0.2844951513 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004453_190 |
0.2855344164 |
- |
- |
- |
| 9 |
Hb_139569_010 |
0.293866375 |
- |
- |
hypothetical protein JCGZ_24635 [Jatropha curcas] |
| 10 |
Hb_006816_160 |
0.3040600444 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 11 |
Hb_012753_180 |
0.3060490072 |
- |
- |
PREDICTED: cytochrome P450 87A3 [Jatropha curcas] |
| 12 |
Hb_000259_100 |
0.3068872263 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 13 |
Hb_000115_260 |
0.3071870141 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH75 [Vitis vinifera] |
| 14 |
Hb_005511_070 |
0.3072639073 |
- |
- |
- |
| 15 |
Hb_005306_200 |
0.3075206004 |
- |
- |
- |
| 16 |
Hb_000903_050 |
0.3077074183 |
- |
- |
hypothetical protein A259_26892 [Pseudomonas syringae pv. actinidiae ICMP 19070] |
| 17 |
Hb_001184_140 |
0.3080256857 |
- |
- |
hypothetical protein B456_008G022200 [Gossypium raimondii] |
| 18 |
Hb_022264_010 |
0.3104665334 |
- |
- |
PREDICTED: uncharacterized protein LOC105795741 [Gossypium raimondii] |
| 19 |
Hb_031022_040 |
0.3104856111 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_041849_010 |
0.3137180075 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |