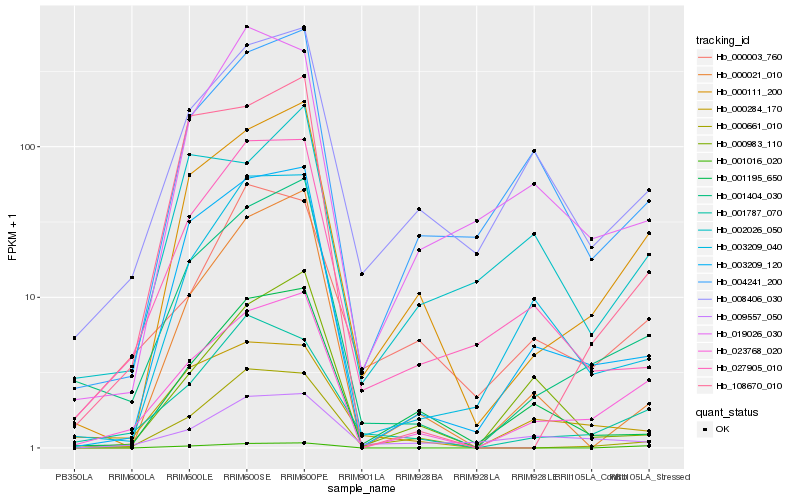
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023768_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001404_030 |
0.1276090498 |
- |
- |
DWNN domain isoform 3 [Theobroma cacao] |
| 3 |
Hb_000111_200 |
0.1309773312 |
- |
- |
PREDICTED: CYSTM1 family protein A-like [Cucumis melo] |
| 4 |
Hb_008406_030 |
0.1679839833 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 5 |
Hb_003209_120 |
0.1687730374 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 6 |
Hb_001787_070 |
0.1802181602 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Camelina sativa] |
| 7 |
Hb_004241_200 |
0.1808780547 |
- |
- |
hypothetical protein JCGZ_00744 [Jatropha curcas] |
| 8 |
Hb_003209_040 |
0.1906223782 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 22-like [Gossypium raimondii] |
| 9 |
Hb_000003_760 |
0.1957490456 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA26-like [Jatropha curcas] |
| 10 |
Hb_001195_650 |
0.1972620216 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 11 |
Hb_027905_010 |
0.2000963198 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 12 |
Hb_000284_170 |
0.2027774006 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_009557_050 |
0.2089624351 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |
| 14 |
Hb_000661_010 |
0.2176852646 |
- |
- |
peroxisomal 3-keto-acyl-CoA thiolase [Hevea brasiliensis] |
| 15 |
Hb_000021_010 |
0.2193650442 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001016_020 |
0.2201400312 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_019026_030 |
0.2208222505 |
- |
- |
hypothetical protein JCGZ_00744 [Jatropha curcas] |
| 18 |
Hb_000983_110 |
0.223440513 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 19 |
Hb_108670_010 |
0.2241921935 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 20 |
Hb_002026_050 |
0.2258697844 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |