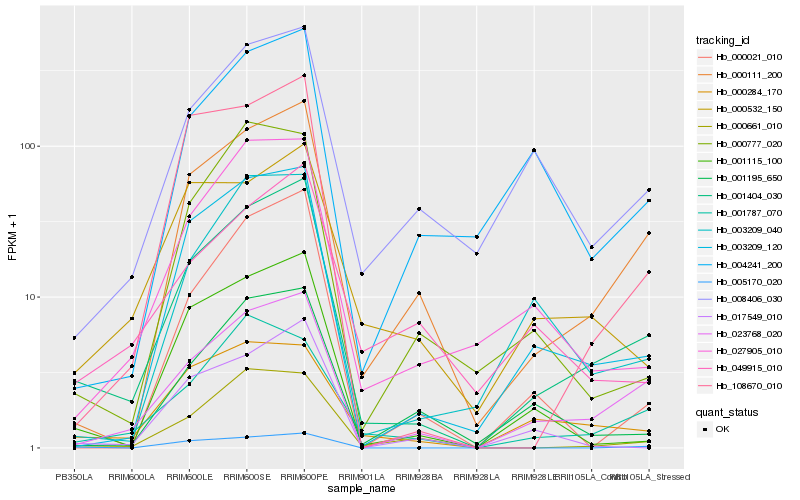
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001404_030 |
0.0 |
- |
- |
DWNN domain isoform 3 [Theobroma cacao] |
| 2 |
Hb_023768_020 |
0.1276090498 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000111_200 |
0.1329181583 |
- |
- |
PREDICTED: CYSTM1 family protein A-like [Cucumis melo] |
| 4 |
Hb_003209_120 |
0.1441436702 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 5 |
Hb_001115_100 |
0.1692822929 |
- |
- |
PREDICTED: uncharacterized protein LOC103714121 [Phoenix dactylifera] |
| 6 |
Hb_005170_020 |
0.1717324127 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 7 |
Hb_027905_010 |
0.1739282162 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 8 |
Hb_108670_010 |
0.1769996488 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 9 |
Hb_000661_010 |
0.1793373252 |
- |
- |
peroxisomal 3-keto-acyl-CoA thiolase [Hevea brasiliensis] |
| 10 |
Hb_000021_010 |
0.1809845118 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003209_040 |
0.1833235214 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 22-like [Gossypium raimondii] |
| 12 |
Hb_008406_030 |
0.1853133922 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 13 |
Hb_000284_170 |
0.1857281517 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_001195_650 |
0.1864647345 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 15 |
Hb_017549_010 |
0.189424612 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 16 |
Hb_001787_070 |
0.1911563163 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Camelina sativa] |
| 17 |
Hb_000532_150 |
0.1962737627 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 18 |
Hb_004241_200 |
0.1978742647 |
- |
- |
hypothetical protein JCGZ_00744 [Jatropha curcas] |
| 19 |
Hb_000777_020 |
0.1983242009 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11 [Jatropha curcas] |
| 20 |
Hb_049915_010 |
0.1988983346 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13 [Jatropha curcas] |