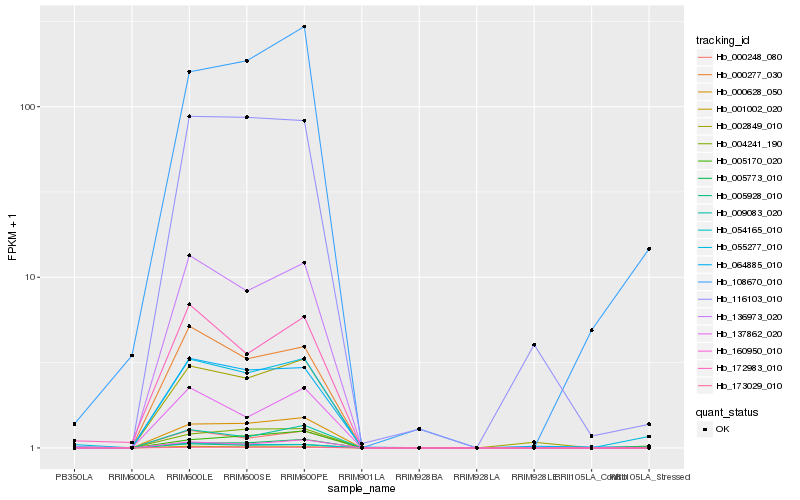
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_055277_010 |
0.0 |
- |
- |
PREDICTED: coenzyme Q-binding protein COQ10 homolog, mitochondrial [Jatropha curcas] |
| 2 |
Hb_002849_010 |
0.1338687354 |
- |
- |
PREDICTED: putative UPF0481 protein At3g02645 [Jatropha curcas] |
| 3 |
Hb_136973_020 |
0.1345816164 |
- |
- |
- |
| 4 |
Hb_005928_010 |
0.1349081667 |
- |
- |
PREDICTED: phospholipase A1-Igamma1, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_108670_010 |
0.137115405 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 6 |
Hb_064885_010 |
0.1385033017 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 7 |
Hb_000628_050 |
0.1410097388 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_001002_020 |
0.1422660265 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_000248_080 |
0.142869181 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 10 |
Hb_000277_030 |
0.146994635 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 11 |
Hb_173029_010 |
0.1471560667 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_054165_010 |
0.1490446445 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 13 |
Hb_116103_010 |
0.1521108651 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 5-like [Jatropha curcas] |
| 14 |
Hb_009083_020 |
0.1527779359 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 15 |
Hb_172983_010 |
0.1541385243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_160950_010 |
0.1541753447 |
- |
- |
PREDICTED: uncharacterized protein LOC105761043 [Gossypium raimondii] |
| 17 |
Hb_005170_020 |
0.1554861305 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 18 |
Hb_005773_010 |
0.1632581006 |
- |
- |
- |
| 19 |
Hb_004241_190 |
0.1645184153 |
- |
- |
- |
| 20 |
Hb_137862_020 |
0.1663208296 |
- |
- |
Leucine-rich repeat receptor protein kinase EXS precursor, putative [Ricinus communis] |