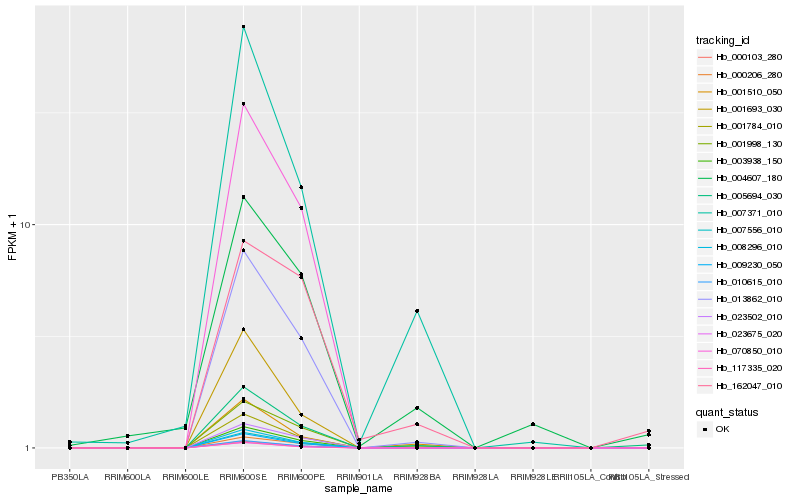
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005694_030 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase PUB22-like [Jatropha curcas] |
| 2 |
Hb_001998_130 |
0.1252097919 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 35 family protein [Populus trichocarpa] |
| 3 |
Hb_013862_010 |
0.1281724443 |
- |
- |
PREDICTED: uncharacterized protein LOC105650642 [Jatropha curcas] |
| 4 |
Hb_001510_050 |
0.1493544475 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_007371_010 |
0.1503486355 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT-like [Populus euphratica] |
| 6 |
Hb_009230_050 |
0.1553531552 |
- |
- |
PREDICTED: transcription factor bHLH113-like [Jatropha curcas] |
| 7 |
Hb_008296_010 |
0.1555477022 |
- |
- |
- |
| 8 |
Hb_001784_010 |
0.1556332641 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 9 |
Hb_010615_010 |
0.156070879 |
- |
- |
PREDICTED: reticuline oxidase-like protein [Jatropha curcas] |
| 10 |
Hb_070850_010 |
0.1564367502 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 1B-like [Jatropha curcas] |
| 11 |
Hb_007556_010 |
0.1565608942 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X2 [Populus euphratica] |
| 12 |
Hb_000103_280 |
0.1566719878 |
transcription factor |
TF Family: HSF |
Heat shock factor protein HSF30, putative [Ricinus communis] |
| 13 |
Hb_003938_150 |
0.1567717421 |
- |
- |
hypothetical protein JCGZ_17711 [Jatropha curcas] |
| 14 |
Hb_001693_030 |
0.161537481 |
- |
- |
PREDICTED: reticuline oxidase-like [Jatropha curcas] |
| 15 |
Hb_004607_180 |
0.1616642344 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_023675_020 |
0.1616645152 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 17 |
Hb_023502_010 |
0.1646927307 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 18 |
Hb_000206_280 |
0.1656081319 |
- |
- |
Uncharacterized protein TCM_031897 [Theobroma cacao] |
| 19 |
Hb_162047_010 |
0.1656743235 |
- |
- |
PREDICTED: protein indeterminate-domain 16 [Jatropha curcas] |
| 20 |
Hb_117335_020 |
0.1674443834 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |