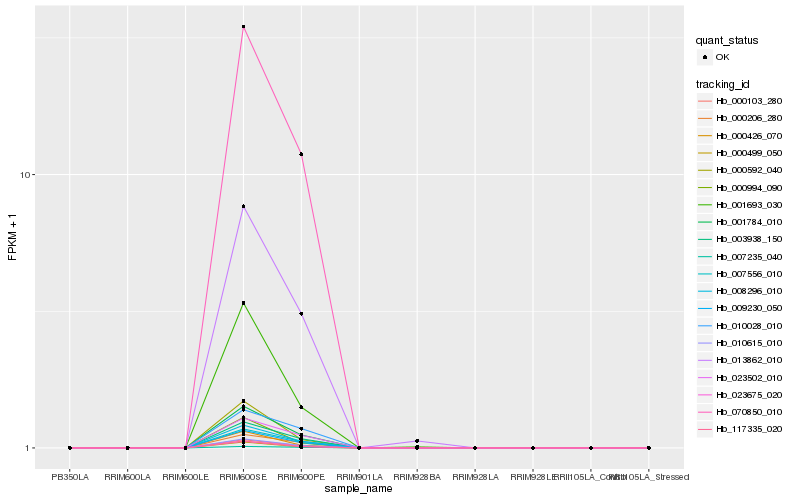
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009230_050 |
0.0 |
- |
- |
PREDICTED: transcription factor bHLH113-like [Jatropha curcas] |
| 2 |
Hb_008296_010 |
0.0054046223 |
- |
- |
- |
| 3 |
Hb_001784_010 |
0.0068907303 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 4 |
Hb_010615_010 |
0.0124028622 |
- |
- |
PREDICTED: reticuline oxidase-like protein [Jatropha curcas] |
| 5 |
Hb_000103_280 |
0.0177250355 |
transcription factor |
TF Family: HSF |
Heat shock factor protein HSF30, putative [Ricinus communis] |
| 6 |
Hb_070850_010 |
0.0218869964 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 1B-like [Jatropha curcas] |
| 7 |
Hb_007556_010 |
0.0229152531 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X2 [Populus euphratica] |
| 8 |
Hb_003938_150 |
0.024551058 |
- |
- |
hypothetical protein JCGZ_17711 [Jatropha curcas] |
| 9 |
Hb_023675_020 |
0.0423425949 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 10 |
Hb_023502_010 |
0.0584746841 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 11 |
Hb_117335_020 |
0.0602662947 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 12 |
Hb_000206_280 |
0.0612037219 |
- |
- |
Uncharacterized protein TCM_031897 [Theobroma cacao] |
| 13 |
Hb_013862_010 |
0.0614695859 |
- |
- |
PREDICTED: uncharacterized protein LOC105650642 [Jatropha curcas] |
| 14 |
Hb_007235_040 |
0.0886871004 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 15 |
Hb_000499_050 |
0.0916826659 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 16 |
Hb_001693_030 |
0.0918649719 |
- |
- |
PREDICTED: reticuline oxidase-like [Jatropha curcas] |
| 17 |
Hb_010028_010 |
0.0920618783 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 18 |
Hb_000592_040 |
0.101943856 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000426_070 |
0.1027912905 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 20 |
Hb_000994_090 |
0.1074389083 |
- |
- |
PREDICTED: putative indole-3-acetic acid-amido synthetase GH3.9 [Jatropha curcas] |